Figure 3.
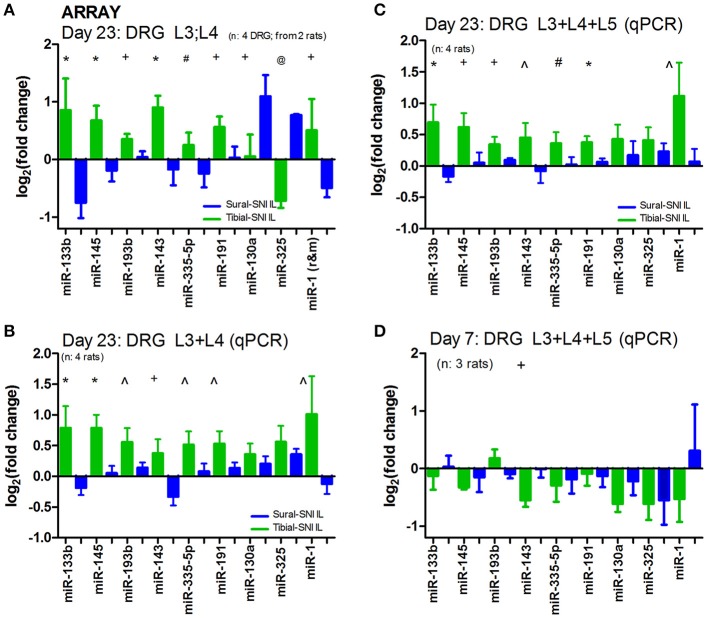
In ipsilateral DRG seven miRs are differentially regulated between Sural-SNI and Tibial-SNI. (A) By using a microarray chip containing probes for detecting 375 miRs, we found that only nine miRs were differentially regulated in IL DRG between Sural-SNI and Tibial-SNI at Day 23. Ct values were <30.60 for miR-133b, <27.78 for miR-145, <27.66 for miR-193b, <30.19 for miR-143, <33.69 for miR-335, <23.42 for miR-191, <37.89 for miR-130a, <36.00 for miR-325, <32.04 for miR-1. Reference miRs were: MammU6-4395470 (Ct values < 19.30), snoRNA135-4380912 (Ct values < 33.98), and U87-4386735 (Ct values < 28.32). The n value was four DRG (two L3 and two L4). For each SNI-group, the mean value from four individual DRG (two L3-DRG and two L4 DRG) were used. (B,C) By using qPCR it was confirmed that seven out of the initially identified nine miRs (in A) were differentially regulated in IL DRG between Sural-SNI and Tibial SNI at Day 23, whether using the data from L3 and L4 (B) or the data from L3, L4, and L5 (C); n = 4 rats. (D) By using qPCR the levels of the identified miRs were measured at Day 7; n = 3 rats. (B–D) The Ct values were <29.94 for miR-133b, <24.67 for miR-145, <27.42 for miR-193b, <34.17 for miR-143, <30.74 for miR-335, < 23.90 for miR-191, <32.17 for miR-130a, <32.62 for miR-325, <30.61 for miR-1. Reference miRs were: snoRNA135-4380912 (Ct values < 30.94) and U87-4386735 (Ct values < 25.16). For each rat, the mean value between their DRG (L3, L4, and L5) was used. In (A–D), the “fold change” (2−ΔΔCT) was obtained by comparing the experimental sample (Sural-SNI or Tibial-SNI ΔCT) vs. the sham sample (Sham ΔCT). Significance between the Tibial-SNI and Sural-SNI: @P < 0.005, *P < 0.02, +P < 0.05, ∧P < 0.07; #P < 0.09, Unpaired t-test, one-tailed. Hence the Sham value will be set to “0” (log2(fold change)=0) (in this and in Figures 4, 5).