Figure 1.
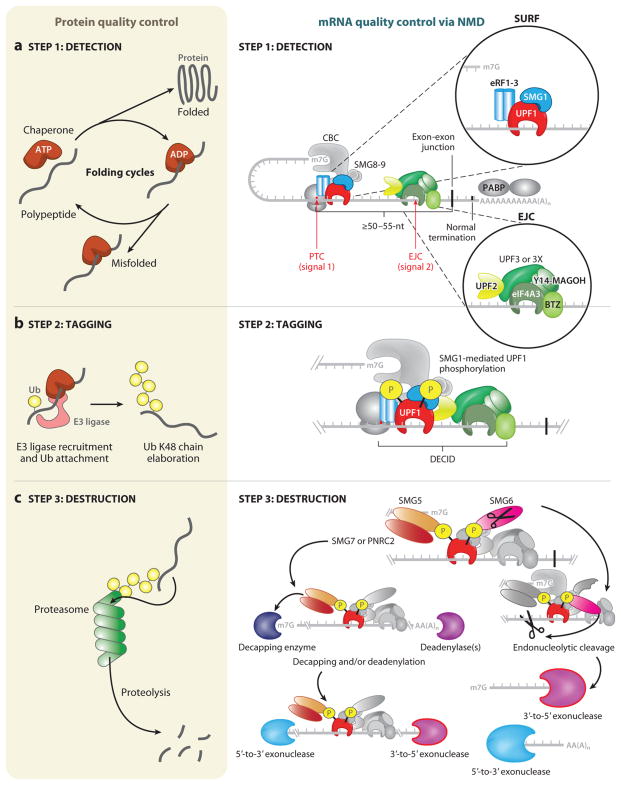
Similar steps govern protein quality control and nonsense-mediated mRNA decay (NMD). A general overview of protein quality control (left) and mRNA quality control, as exemplified by NMD (right), can be separated into three distinct steps: detection, tagging, and destruction. (a) Detection for protein quality control proceeds through ATP-dependent cycles of chaperone binding, release, and rebinding to hydrophobic patches in unfolded and/or partially folded proteins, providing the opportunity to fold (left). A decision is made as to whether the client protein is terminally misfolded, and, if so, this leads to tagging. The detection step for NMD relies on two signals (right). The first signal is provided by the premature termination codon (PTC), which generally defines NMD substrates, and the proteins that associate with the translation termination complex that assembles at the PTC, including the terminating ribosome and the SURF complex (inset, upper right), which consists of SMG1, UPF1, eRF1, and eRF3. The second signal often derives from an exon-junction complex (EJC), which associates with mRNAs <24 nucleotides (nts) upstream of a splicing-generated exon-exon junction whether or not they are NMD substrates. Other signals, such as unusually long 3′ UTRs, also exist. The EJC (inset, lower right) is composed of four core components (eIF4A3, Y14, MAGOH, BTZ) and associated NMD factors UPF3 or UPF3X and UPF2. If a PTC is located ≥50–55 nts upstream of an exon-exon junction, the mRNA is recognized as aberrant and tagging proceeds. (b) (Left) Tagging for misfolded proteins is governed by the ubiquitylation system. In an ATP-dependent reaction, ubiquitin (Ub) is covalently transferred from an E1 enzyme to an E2 enzyme (not shown). Ub can be transferred in covalent linkage to E3 enzymes containing a HECT (homologous to the E6-AP carboxyl terminus) domain before transfer to the substrate protein, or Ub can be directly transferred from the E2 enzyme to substrates by E3 ligase enzymes containing a RING (really interesting new gene) domain. E3 ligases can mediate ubiquitin transfer to misfolded proteins by binding to molecular chaperones. Mono-Ub is elaborated into a Ub chain with linkages at lysine (K)48. This constitutes the tag that identifies the attached protein for destruction. (Right) For tagging during NMD to occur, all or some of the SURF complex joins the EJC, possibly while the terminating ribosome is still present, forming a decay-inducing complex (DECID). This configuration activates SMG1 to phosphorylate UPF1. Phosphorylated UPF1 signals the mRNA for destruction and has the added effect of inducing translational repression of the mRNA, which is a prerequisite for destruction. (c) (Left) Destruction of proteins is the job of the proteasome, a macromolecular proteolytic machine whose components include proteins that recognize Ub-chain tags and initiate feeding of the aberrant protein into the bore of the proteasome. Aberrant proteins are thereby degraded into short peptides. (Right) NMD-dependent destruction of mRNA relies on recruitment of SMG6 and/or SMG5-SMG7 or SMG5 -PNRC2 complexes via the phosphate tags on UPF1. SMG6 possesses its own endonucleolytic cleavage activity, cutting the mRNA target 5′ to the EJC. 3′-to-5′ exonucleases degrade the 5′-cleavage product. UPF1 helicase activity disassembles the RNP components bound to the 3′-cleavage product, and this is followed by 5′-to-3′ exonucleolytic degradation. The SMG5-SMG7 or SMG5-PNRC2 adaptor complexes, recruited via SMG5 to a UPF1-localized phosphate moiety, direct exonucleolytic degradation of the mRNA. These adaptors recruit decapping enzymes and/or deadenylation enzymes, and their activities are followed by 5′-to-3′ and 3′-to-5′ exonucleolytic decay, respectively. Proteins relevant to each step are shown in color. Abbreviation: CBC, cap-binding complex.