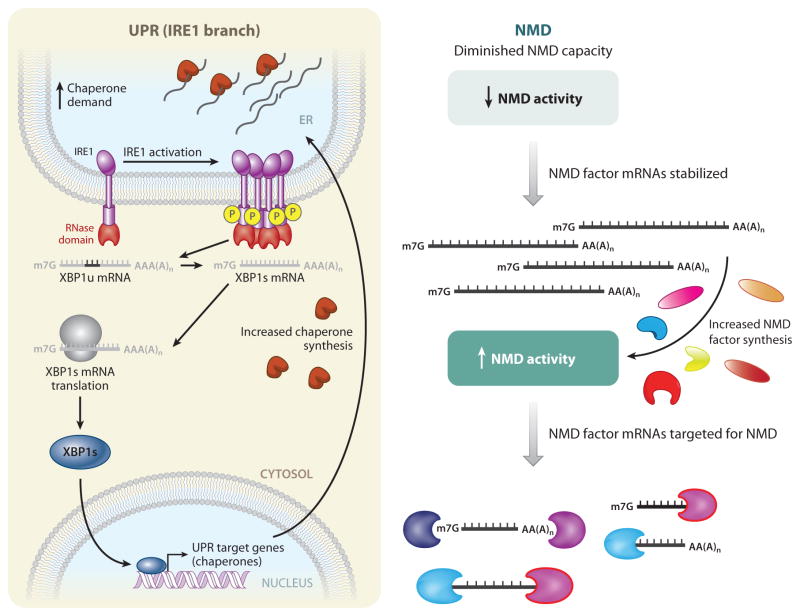
Figure 2.
Regulation of protein quality control and nonsense-mediated mRNA decay (NMD) provides buffering capacity. Protein quality control (left), as exemplified by the IRE1-mediated branch of the unfolded protein response (UPR), and mRNA quality control, as exemplified by NMD (right), are both governed by regulatory loops that provide buffering capacity. (Left) For IRE1-mediated UPR activation, increased demand on the protein-folding load of the endoplasmic reticulum (ER) leads to an accumulation of unfolded and/or partially folded proteins and an increased demand for more folding chaperones. IRE1, a membrane-embedded sensor of unfolded proteins, is activated either by direct binding to unfolded proteins or by titration of chaperones (which prevent IRE1 oligomerization) away from IRE1. IRE1 subsequently oligomerizes and autophosphorylates. The RNase domain in activated IRE1 (red ) then mediates an unconventional cytoplasmic splicing reaction to generate spliced XBP1 (XBP1s) mRNA from unspliced (XBP1u) mRNA. XBP1s mRNA is translated into XBP1s protein, which is a transcriptional activator that upregulates a suite of UPR target genes. Among these genes are chaperones, leading to increased chaperone synthesis so as to match the protein-folding capacity of the ER with cellular demand. (Right) Some NMD factors (UPF1, UPF2, UPF3X, SMG1, SMG5, SMG6, and SMG7) are themselves normally targets of NMD by virtue of the presence of an unusually long 3′ UTR, a uORF, or both. When genetic insults diminish NMD capacity, NMD activity is decreased, and the mRNAs encoding the NMD factors are stabilized. These stabilized mRNAs direct protein synthesis in an effort to restore normal NMD activity.