FIGURE 7.
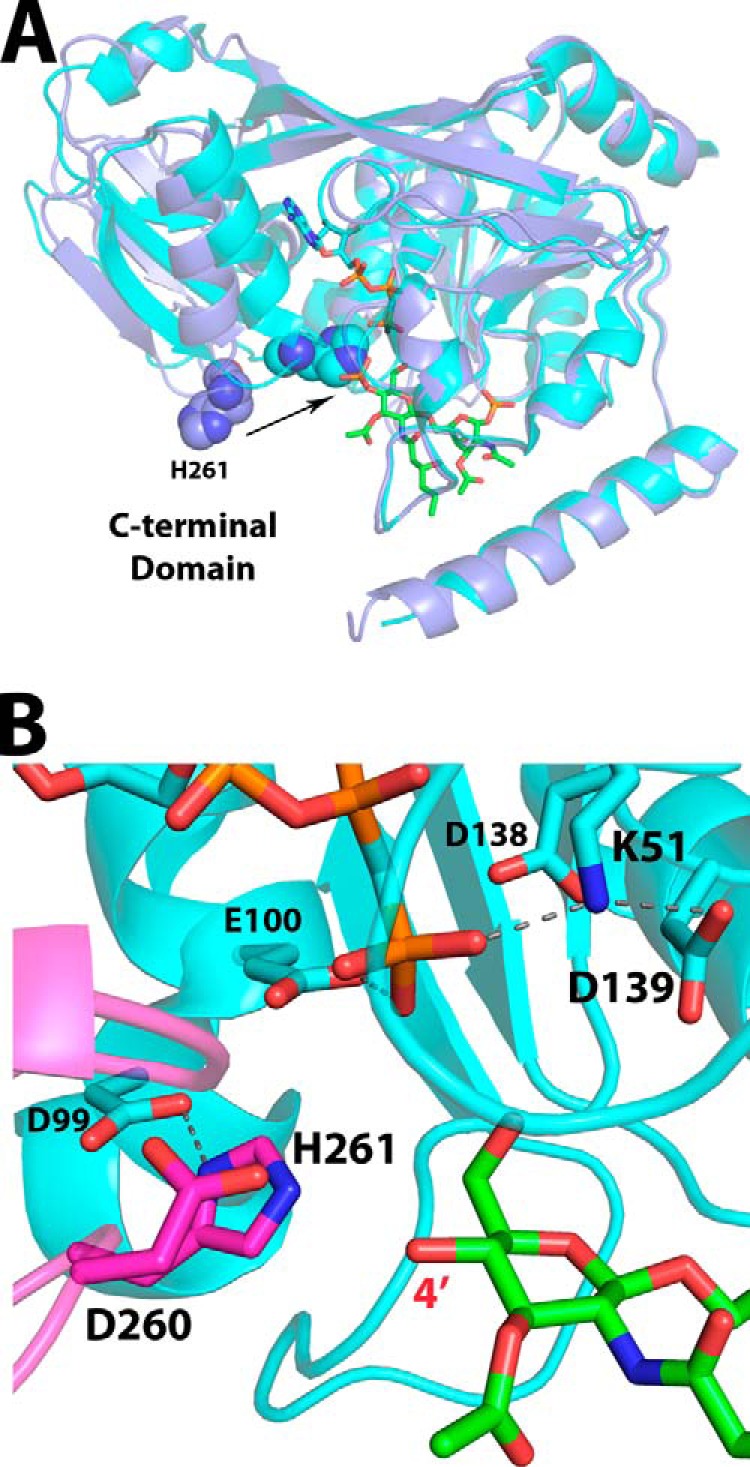
Overlay of AMP-PCP and lipid IVA-bound structures as a model for the catalytic ATP-DSMP-LpxK ternary complex. A, alignment of the LpxK-lipid IVA complex (purple schematic) and the nucleotide-bound complex in the “closed” form (Protein Data Bank code 4ITL) (cyan schematic) illustrates how the motion of the C-terminal domain closing around the nucleotide assembles the active site. The movement of the catalytic histidine His261 (spheres) upon closure of the C-terminal domain to assemble the active site is indicated by an arrow. B, visualization of the active site of the AMP-PCP bound LpxK structure (cyan schematic and sticks) with DSMP modeled (green sticks, with atoms for the 4′-phosphate removed) based on the alignment of the N-terminal domains from A. The C-terminal domain (magenta) closure moves the α-carbon of His261, the putative catalytic base, over 7 Å so that it may interact with Asp99 to be activated. Among the plausible catalytic bases Glu100, Asp139, and His261, the histidine resides nearest to the 4′-hydroxyl position (red) at 3.3 Å in this model, giving further credence to the hypothesis that this residue is the catalytic base. Hydrogen bonds within the AMP-PCP-bound LpxK structure are indicated with gray dashes. Font size is indicative of depth.
