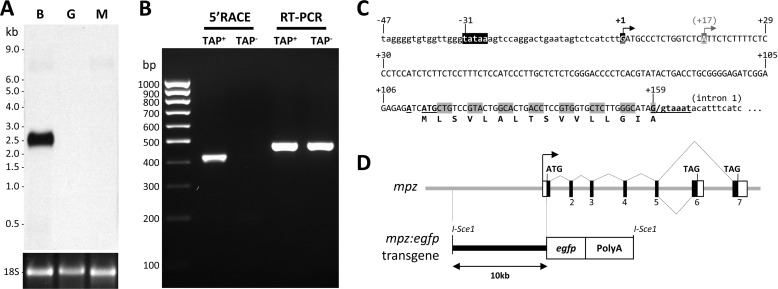
FIGURE 1.
mpz transcript and promoter. A, a Northern blot of total RNA (3 μg/lane) from brain (B), muscle (M), and gut (G) was probed using a cRNA probe to the open reading frame of mpz shared by all known splice variants. The ethidium bromide-stained 18 S rRNA loading control is shown below the blot, and molecular size markers are annotated to the left. B, 5′-RACE was used to map the transcriptional start site of mpz. The picture shows an ethidium bromide-stained agarose gel under UV illumination following electrophoretic separation of PCR products. 5′-RACE (lanes 2 and 3) and RT-PCR (lanes 4 and 5) were carried out using cDNA derived from total brain RNA that was either pretreated with tobacco acid pyrophosphatase (TAP+; lanes 2 and 4) or received no treatment (TAP−; lanes 3 and 5) prior to adapter ligation. C, the promoter region of mpz is shown. The arrows indicate the major and minor start sites at positions +1 and +17, respectively. The consensus TATA box at position −31 is highlighted in black. The open reading frame of exon 1 is indicated. The consensus translational initiation sequence at position +114 and splice donor consensus at position +159 are underlined and in boldface type. D, the diagram depicts the genomic organization and alternative splicing of mpz. Exons are numbered as previously reported (17). A schematic map of the 10-kb mpz:egfp construct is shown below the genomic map.