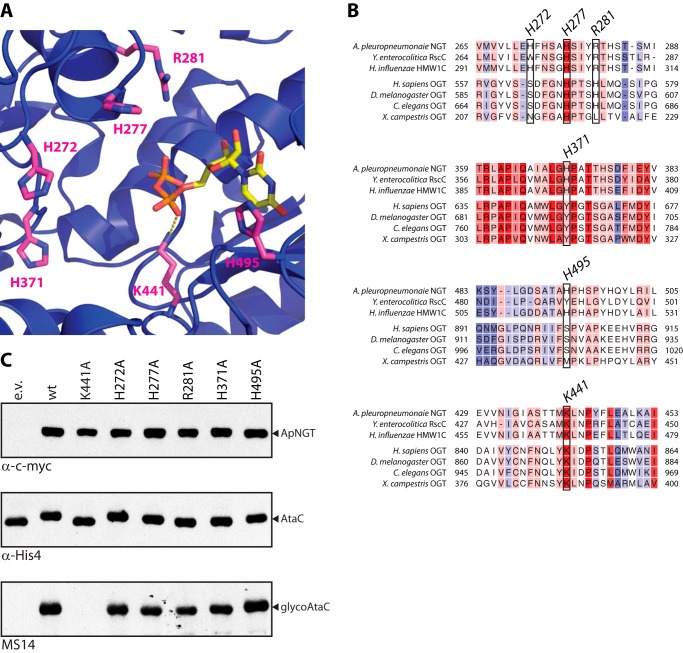
FIGURE 5.
Basic amino acids around the active site of APNGT are not involved in catalysis. A, crystal structure of ApNGT in complex with UDP (PBD code 3Q3H) (22). All basic residues in an 8-Å radius from the β-phosphate of the UDP moiety (in yellow) are marked in magenta. B, structure-aided sequence alignments of several GT41 glycosyltransferase sequences. The region around the residues marked in A is shown. For full alignment see supplemental Fig. S2. Alignment was performed using Expresso (28). Coloring indicates conservation of residues (from blue, not conserved, to red, fully conserved). C, in vivo activity assay of ApNGT mutants. The different mutants of ApNGT were co-expressed in E. coli with acceptor substrate AtaC1866–2428 and whole cell protein extracts were analyzed by immunoblot. NGT proteins were detected via the Myc epitope (top panel), AtaC1866–2428 via the His10 tag (middle panel), and glycosylation was detected using Asn-Glc specific serum MS14 (bottom panel) (23, 32).