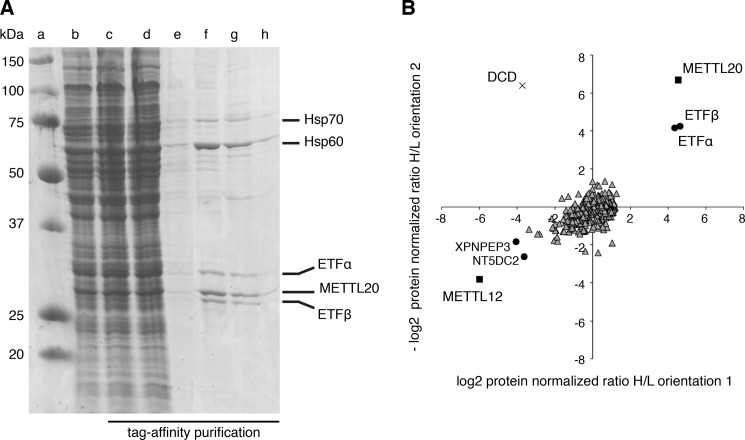
FIGURE 2.
Identification of proteins associated with METTL20. A, SDS-PAGE analysis of METTL20 and associated proteins purified by affinity chromatography from Flp-In T-REx HEK293T cells expressing FLAG-StrepII-tagged METTL20. Lane a, molecular mass marker proteins (molecular masses indicated on the left); lane b, solubilized mitochondria; lane c, unbound proteins; lanes d and e, first and final column washes; lanes f–h, eluates 1, 3, and 5; on the right of the gel are shown the identities of the stained bands determined by tandem-MS analysis of tryptic peptides from the bands in lanes f and g. B, quantitative mass spectrometry of METTL20 and associated proteins. SILAC-labeled cells from METTL20 and differentially labeled control cells overexpressing METTL12 were combined and purified as above. The experiment was performed in both labeling orientations (orientation 1, METTL20-heavy plus METTL12-light; orientation 2, METTL20-light plus METTL12-heavy). Each data point corresponds to the abundance ratio of an identified protein from the two complementary experiments. ■, METTL20 and METTL12; ●, proteins significantly associated with METTL20 or METTL12 in both orientations; ▵, 721 proteins that comprise 709 insignificantly associated with either tagged-protein (including HSP70) and 12 proteins with one significant orientation only, including HSP60; ×, exogenous contaminant (DCD, dermcidin).