FIGURE 2.
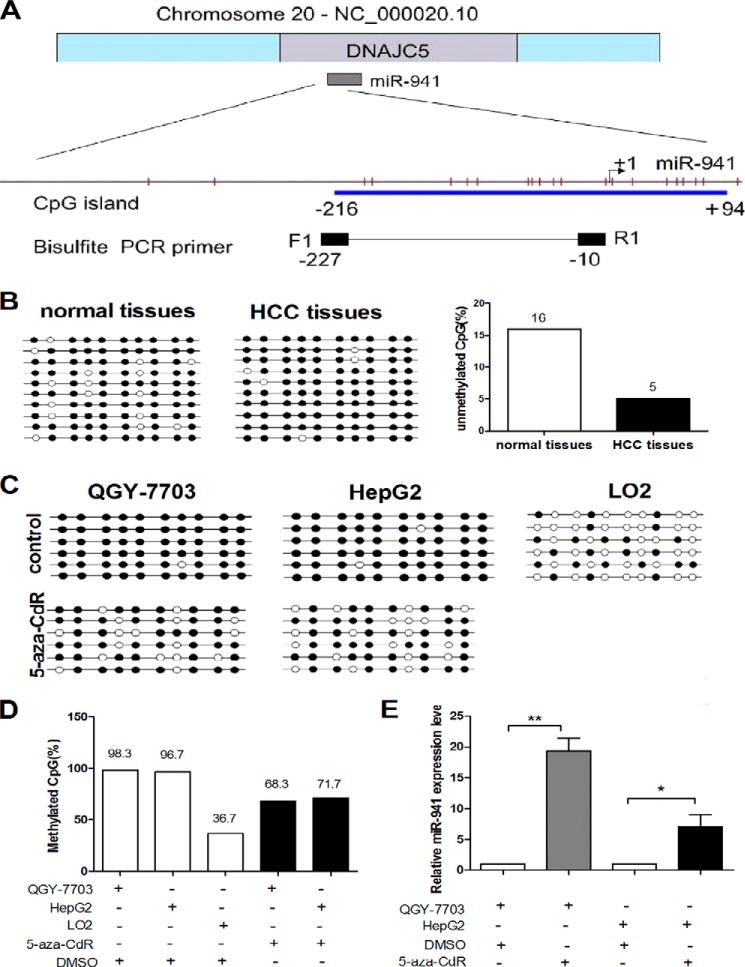
miR-941 is regulated by DNA methylation in QGY-7703 and HepG2 cell lines. A, genomic representation of Chr20, which includes the DNAJC5 gene and miR-941 is embedded in intron-1 of DNAJC5. Relative location of CpG sites (the red vertical bars) are indicated upstream of miR-941 and the start site is designated as +1. The CpG island (blue box) and primers for bisulfite PCR (black box) are also indicated. B, we carried out a bisulfite sequencing analysis to determine the DNA methylation status of each of the 10 CpGs in HCC tumor tissues and their adjacent non-tumor tissues. Open and filled squares denote unmethylated and methylated CpG sites, respectively. Each row represents a single clone. C, the methylation status of the CpG island is assessed before and after 5-aza-CdR treatment in HCC cell lines or LO2 cells. D, the data in C was presented as percentages of the methylated/total CpGs in the plot. E, qRT-PCR detected the level of miR-941 expression in QGY-7703 and HepG2 cell lines after treatment with 5-aza-CdR compared with the control of treatment with DMSO (*, p < 0.05; **, p < 0.01).