Figure 4.
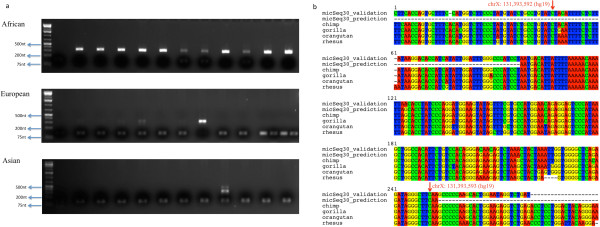
PCR of individuals and evolutionary conservation of micSeq30. a) PCR results for micSeq30 for samples from three populations used for validation. The allele frequency clearly shows population stratification for this micSeq: with higher frequency in African, and much lower in others, p-value < 10−5 for African vs non-African (Table 1). b) Sanger sequencing results for PCR amplified DNA, predicted sequence of micSeq30, and homologs from other primates. The genomic coordinates for other primates are: chimp: chrX: 132, 861, 450-132, 861, 750; gorilla: chrX: 129, 774, 750-129, 775, 050; orangutan: chrX: 131, 684, 602-131, 684,902; and rhesus: chrX: 130, 465, 777-130, 466, 071. The red arrows show the corresponding coordinates in human reference genome (hg19). The inserted position of micSeq30 in the reference genome is: chrX: 131, 393, 592, and, the corresponding anchor reads for micSeq30 locate in the region of chrX: 131, 393, 154-131, 393, 947.
