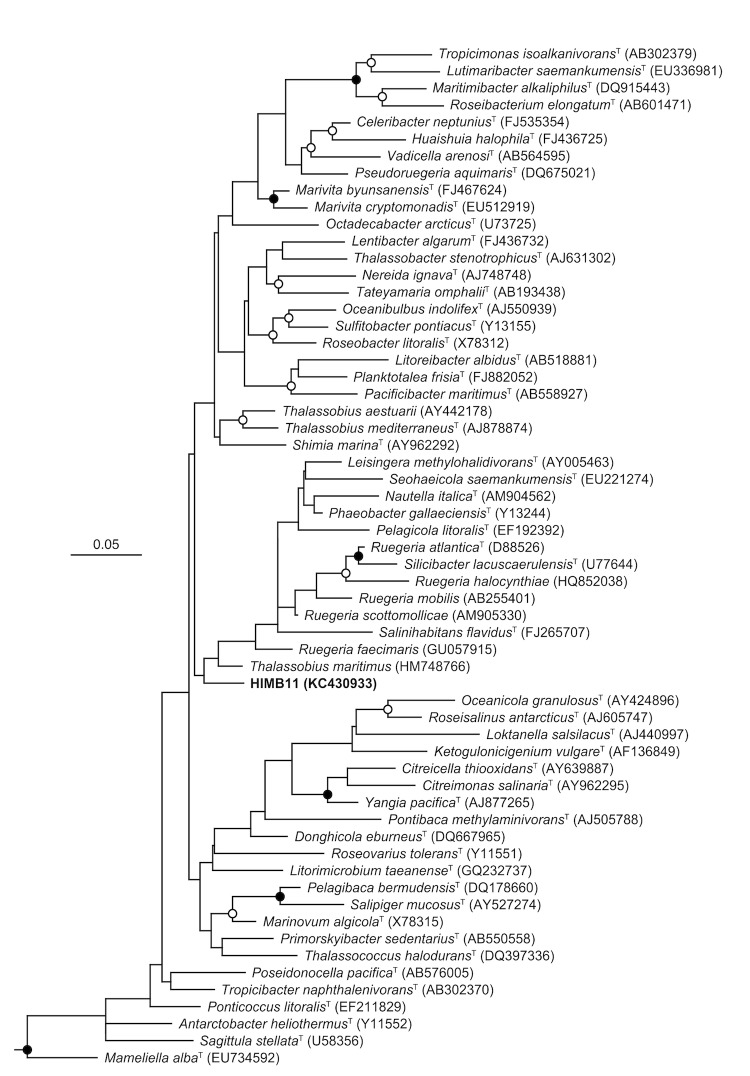
Figure 1.
Phylogenetic relationships between HIMB11 and bacterial strains belonging to the Roseobacter clade. SSU rRNA gene sequences were aligned with version 111 of the ‘All-Species Living Tree’ project SSU rRNA gene database [16] using the ARB software package [17]. The phylogeny was constructed from nearly full-length gene sequences using the RAxML maximum likelihood method [18] within ARB, filtered to exclude alignment positions that contained gaps or ambiguous nucleotides in any of the sequences included in the tree. Bootstrap analyses were determined by RAxML [19] via the raxmlGUI graphical front end [20]. The scale bar corresponds to 0.05 substitutions per nucleotide position. Open circles indicate nodes with bootstrap support between 50-80%, while closed circles indicate bootstrap support >80%, from 500 replicates. A variety of Archaea were used as outgroups.