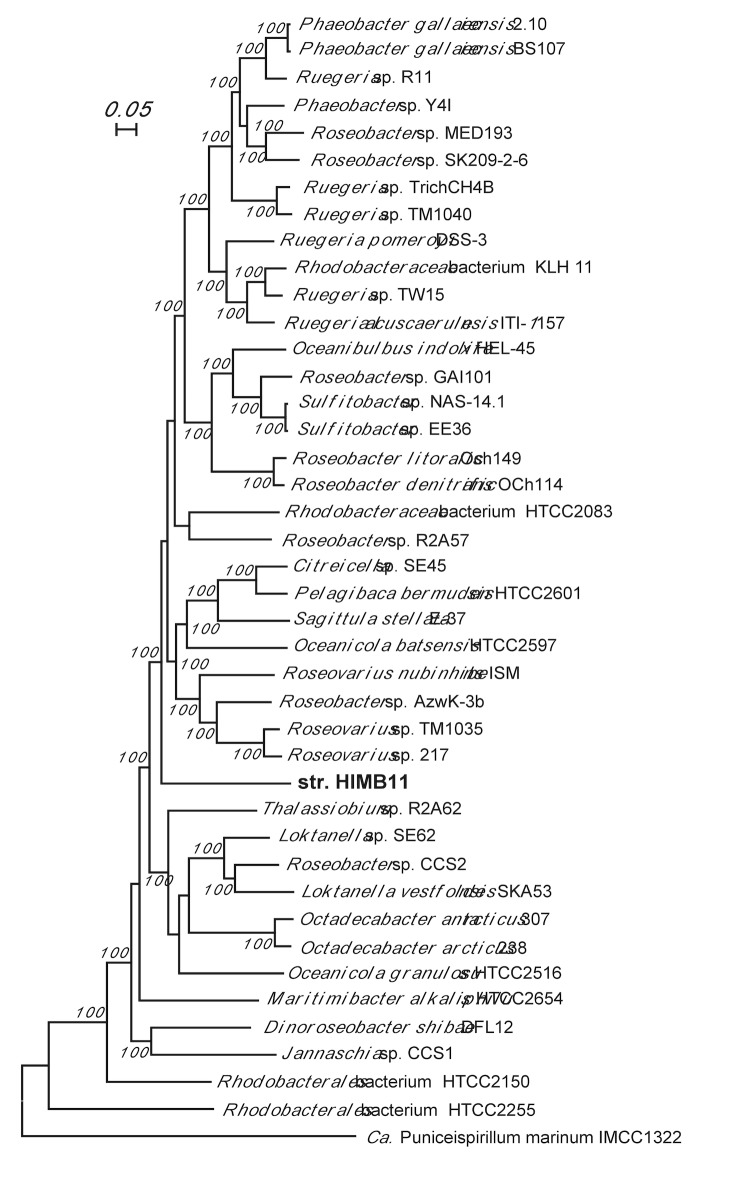
Figure 2.
Maximum likelihood phylogenomic tree derived from a concatenation of 719 shared single-copy orthologous protein sequences from strain HIMB11, 40 other Roseobacter members, and the SAR116 clade member Ca. Puniceispirillum marinum IMCC1322 (outgroup) using the RAxML maximum likelihood method [18]. After a reciprocal BLAST search [22] was performed for all possible genome pairs at the amino acid level using an E value of 10-6, orthologous genes in each of the genome pairs were identified using the MSOAR software [23], which assigns orthologs by considering both sequence similarity and gene context information. One genome was picked at random as the reference genome, and pairwise orthologs were linked to the reference. Values at the nodes show the number of times the clade defined by that node appeared in the 100 bootstrapped data sets. The scale bar indicates number of substitutions per site. Bootstrap values lower than 50% were not displayed.