Figure 4. Fendrr binds to the PRC2 and TrxG/MLL complexes and to target promoters.
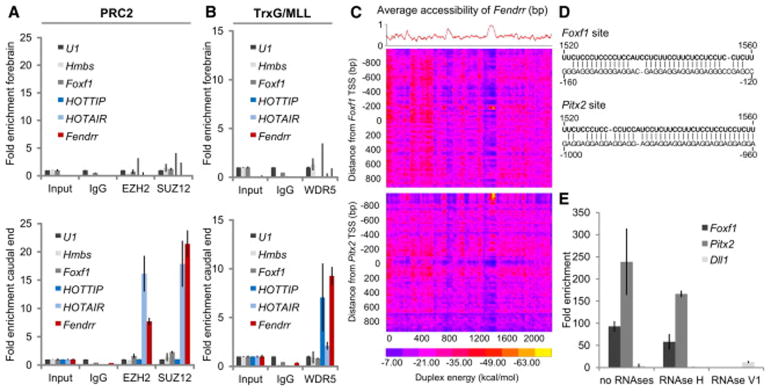
(A, B) RNA co-immunoprecipitation (RIP) from forebrain (upper panels) and caudal end (lower panels) lysates from wild type embryos using antibodies directed against the PRC2 components EZH2 and SUZ12 (A) or the TrxG/MLL component WDR5 (B); normal rabbit IgG was used as a control. Fold enrichment has been normalized to non-enriched input sample and U1 rRNA. Fendrr transcripts co-precipitated with EZH2, SUZ12, and WDR5 from caudal end tissue only, while the control lncRNA Hotair co-precipitated only with EZH2 and SUZ12 and HOTTIP lncRNA co-precipitated only with WDR5 from the caudal end tissue. Foxf1 and Hmbs RNA co-precipitation was used as a negative control. Mean +/− s.d. are shown (n=3). (C) Binding potential between Fendrr and genomic regions. The red curve shows the average probability of single stranded RNA as computed by sfold with a length parameter of 200 and W=1 (Ding et al. 2004). The heat map represents the base-pairing energy for an RNA/RNA duplex model for 40bp regions along the Fendrr transcript and 2,000 bp around the TSS of Foxf1 (top) and Pitx2 (bottom). The duplex energy is computed for each such region, staggered by 20bp. The optimal base pairing occurs in a region of Fendrr that also strongly favors single-stranded RNA, suggesting an open conformation that would enable the binding to the promoter. Probabilities are then averaged for a sliding window of 40bp to give the average RNA accessibility of the region that is binding. (D) Representation of the predicted interaction of the Fendrr RNA region and the promoter DNA region exhibiting the lowest free energy of approximately −70 kcal/mol (see yellow spot in C). (E) In vitro RNA/dsDNA binding assay utilizing biotin tagged RNA oligos as bait. Bars represent normalized enrichment of indicated 2,000 bp promoter fragment over background using a control RNA oligonucleotide (n=3, Mean +/− s.d. is shown).
