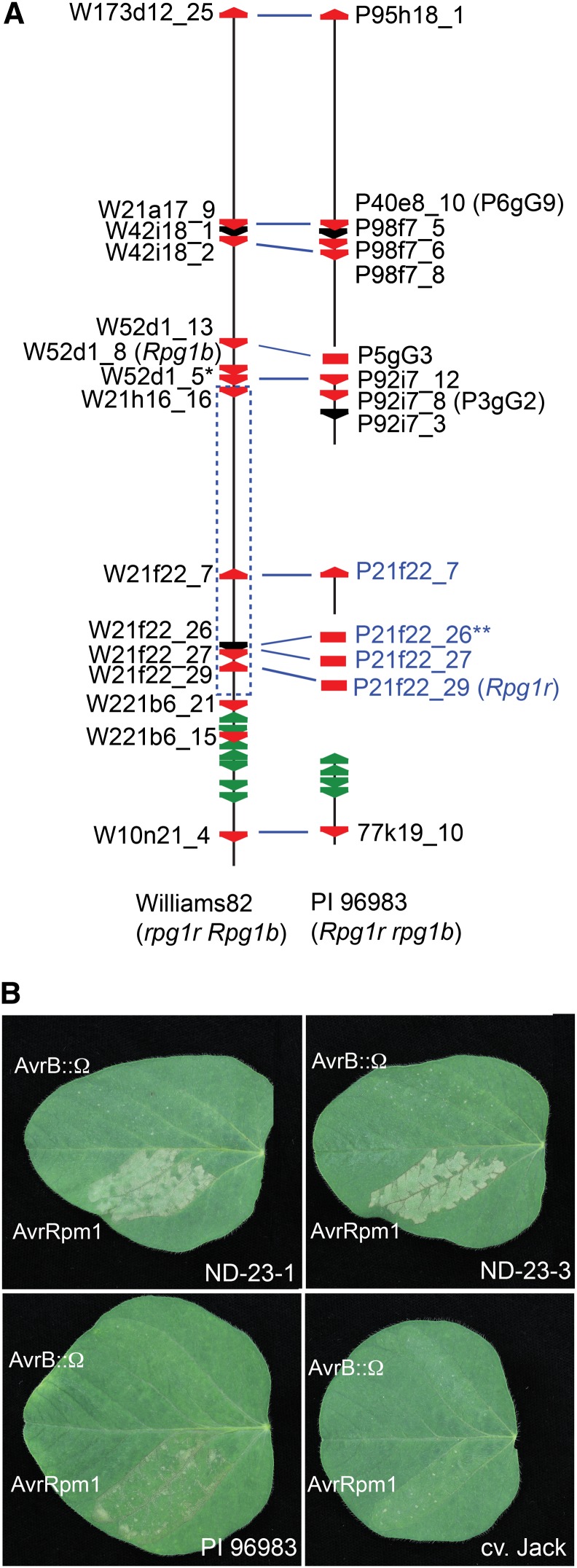
Figure 1.
Cloning of the soybean Rpg1r gene. A, Distribution of NB-LRR genes across an approximately 1-Mb interval encompassing the Rpg1r locus in cv Williams 82 and PI 96983. The vertical black lines represent genomic sequence from soybean cv Williams 82 (rpg1r Rpg1b) and PI 96983 (Rpg1r rpg1b) as described by Innes et al. (2008). The region corresponds to the interval from Glyma13g25440 to Glyma13g26530 in the soybean WGS (Schmutz et al., 2010). Breaks in the lines indicate lack of available sequence. The red and green polygons correspond to CNL and TIR-NB-LRR family members, respectively. Black polygons represent CNL pseudogenes or paralogs that extend beyond the available sequence. Gene names printed in black correspond to CNL genes identified in the available genomic sequence, while those printed in blue correspond to CNL genes amplified from PI 96983 total genomic DNA. The blue dashed rectangle indicates the genetically defined region containing Rpg1r (see Supplemental Fig. S1). The blue lines linking CNL genes in the cv Williams 82 and PI 96983 sequences indicate probable alleles. A single asterisk indicates W52d1_5 has the atypical structure CNL:CNL. Double asterisks indicate gene 21f22_26 is a pseudogene in both cv Williams 82 and PI 96983. B, Functional complementation of Rpg1r recognition specificity in soybean stable transformants. Primary leaves from accession PI 96983 (Rpg1r), untransformed cv Jack (rpg1r), and cv Jack transformants expressing an Rpg1r transgene were injected with PsgR4 expressing avrRpm1 or a nonfunctional avrB allele disrupted with an Ω sequence (avrB::Ω). The responses in two independent transformants (ND-23-1 and ND-23-3) are shown. Photographs were taken approximately 24 h after injection.