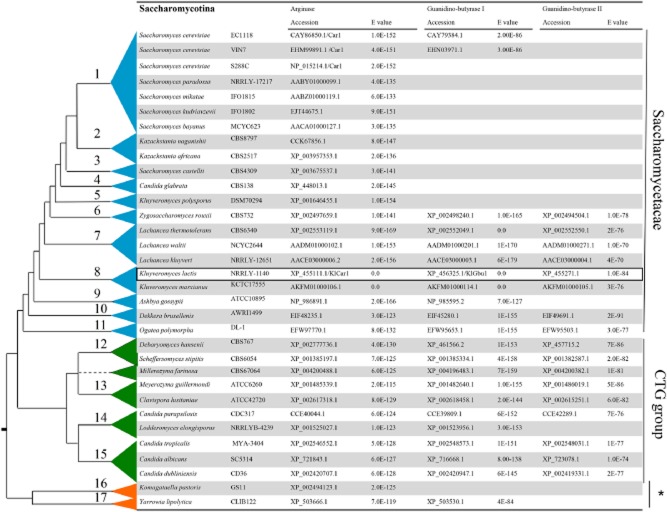
Fig. 7.
Phylogenetic tree of Saccharomycotina yeast species and analysis of the distribution of K. lactis KlGBU1/KLLA0F27995g and KLLA0F04235g orthologues within this subphylum. The tree is based on Kurtzman (2003) and Dujon (2010). K. lactis orthologous amino acid sequences were identified by Blastp or tBlastn in genomics data of each individual species mentioned. The branch length is arbitrary. The reported species are divided in several groups: The subphylum Saccharomycetaceae (in blue) includes the following clades: 1 – Saccharomyces sensu stricto, 2 – Kazachstania, 3 – Naumovozyma, 4 – Nakaseomyces, 5 – Vanderwaltozyma, 6 – Zygosaccharomyces, 7 – Lachancea, 8 – Kluyveromyces, 9 – Eremothecium, 10 – Torulaspora, 11 – Ogataea, The CGT group (in green) includes the clades: 12 – Debaryomyces, 13 – Clavispora, 14 – Candida I, 15 – Candida II; The Dipodascaceae and related families (*) (in orange) includes: 16 – Komagataella, 17 – Yarrowia. Label accession indicates the specific amino acid sequence accession number and E-value indicates the significance of the comparison, the significance threshold was set at 1.0E-70. The K. lactis sequences used as query for the Blast analysis were indicated in a box.