Figure 1. Non-coding EBV RNAs.
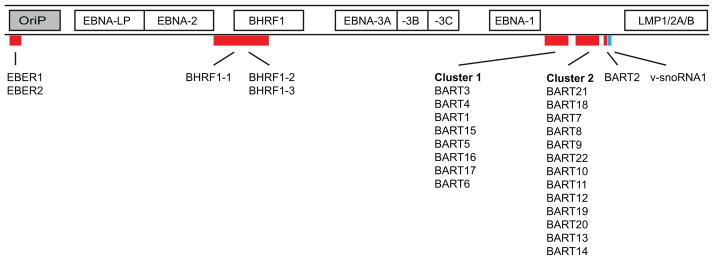
This schematic illustration shows the locations of non-coding RNAs within EBV’s genome. Along with the non-coding nuclear EBER RNAs, and a snoRNA [36], EBV encodes at least 25 pre-miRNAs located within two regions of the genome. The smaller subset of BHRF1 miRNAs is derived from transcripts mapping approximately between 53,000 and 56,000 bps. The majority of EBV miRNAs arise from highly spliced transcripts mapping approximately between 139,000 and 153,000 bps. Also shown are the latent origin of replication (OriP) and genes expressed in latently infected cells. Note: the figure is not drawn to scale and is a linear representation of EBV’s circular genome which encompasses approximately 165,000 bps.
