Abstract
Profiling of protein species is important because gene polymorphisms, splice variations and post-translational modifications may combine and give rise to multiple protein species that have different effects on cellular function. Two-dimensional gel electrophoresis is one of the most robust methods for differential analysis of protein species, but bioinformatic interrogation is challenging because the consequences of changes in the abundance of individual protein species on cell function are unknown and cannot be predicted. We conducted DIGE of soleus muscle from male and female rats artificially selected as either high- or low-capacity runners (HCR and LCR, respectively). In total 696 protein species were resolved and LC–MS/MS identified proteins in 337 spots. Forty protein species were differentially (P < 0.05, FDR < 10%) expressed between HCR and LCR and conditional independence mapping found distinct networks within these data, which brought insight beyond that achieved by functional annotation. Protein disulphide isomerase A3 emerged as a key node segregating with differences in aerobic capacity and unsupervised bibliometric analysis highlighted further links to signal transducer and activator of transcription 3, which were confirmed by western blotting. Thus, conditional independence mapping is a useful technique for interrogating DIGE data that is capable of highlighting latent features.
Keywords: Animal Selection Model, Bibliometric network analysis, Mass spectrometry, N-myc down-regulated gene 2, Sexual dimorphism, Signal transducer and activator of transcription 3
1. Introduction
The statistical link between exercise capacity and all-cause mortality is irrefutable [1] but the mechanisms that underpin the preventive effects of physical activity are not yet fully understood. Previously, we have used proteomic techniques to investigate adaptations in human skeletal muscle [2], and in the heart [3] and skeletal muscle [4] of outbred rats. However, it is difficult to know whether changes measured in the proteome after a period of exercise training reflect an acute response to the last bout of exercise or a summative chronic response to the increase in habitual activity. Moreover, the concurrent acute effects and chronic responses to exercise may be influenced by genetic factors that affect either baseline levels or the responsiveness to training stimuli. To minimise these complications and specifically investigate the link between aerobic capacity and disease risk we have used divergent artificial selection in a genetically heterogeneous population of rats to create two populations with markedly different intrinsic running capacities [5]. Early in the selection process it was noted [6] that low capacity runners (LCR) were insulin resistant and displayed cardiovascular risk factors including hypertension and dyslipidaemia. In contrast, high capacity runners (HCR) exhibit enhanced metabolic flexibility including augmented muscle glucose uptake and oxidation in the presence of insulin, as well as heightened uptake and oxidation of lipids under fasting conditions [7]. These differences in substrate utilisation between HCR and LCR are associated with greater abundance and efficiency of mitochondria [8] as well as enhanced insulin receptor signalling [9] in HCR animals. As such these findings add further associational evidence linking muscle aerobic capacity with metabolic disease risk, but the regulatory networks driving these phenomena remain to be discovered.
Transcriptome profiling of HCR/LCR muscle [10] reported 239 differentially expressed genes, and gene-set enrichment analysis found correlations between free-wheel running capacity and gene-sets including oxidative phosphorylation, fatty acid oxidation and PPAR signalling. Furthermore, label-free LC–MS profiling [11] provided protein-level confirmation for the greater abundance of heart-type fatty acid binding protein, cytochrome b-c1 complex subunit 1 and cytochrome c oxidase subunit 4, isoform 1 in HCR skeletal muscle. Nonetheless, divergence between HCR and LCR populations is likely to involve complex epistatic interactions and additive selection of genes consisting of different alleles, which may create differences at the protein-species level [12]. Proteome mining studies (e.g. [13,14]) have used sophisticated orthogonal separations and LC–MS/MS analysis to catalogue approximately 2000 of the most abundant muscle proteins. However, it is not possible to analyse large numbers of protein species (also known as ‘proteoforms’ [15]) using these techniques. This is because tryptic digestion severs the link between the different combinations of post-translational modifications that make up each species. In contrast, protein species can be readily resolved using 2D gel electrophoresis based on differences in isoelectric point (pI) and relative mass (Mr) [4,16]. Indeed, using 2D difference in-gel electrophoresis (DIGE) we [17] discovered the first evidence that the cardiac proteome of LCR is exposed to greater oxidative stress, based on modification of metabolic enzymes by the cytotoxic aldehyde, 4-hydroxynonenal (4-HNE). Our DIGE analysis resolved almost 1000 cardiac protein species but it was difficult to exploit this highly parallel data using mainstream bioinformatic tools that assume greater abundance of transcripts or proteins can be inferred to mean a greater contribution to cellular function. For example, an increase in the density of a 2Dgel spot may in some cases depict a greater abundance of a relatively inactive species of that protein and hence a decrease in the related cellular function. Because we do not know the entire proteome and are unable to predict the role of each species of a protein, pragmatic methods for finding associations between protein species are needed that do not rely on prior knowledge. In particular, it is important to seek ‘un-biased’ or unsupervised approaches because our intuitive interpretations are not objective and are influenced strongly by our perception of which proteins/events/pathways etc. are of greatest importance in particular scenarios.
Proteins interact with each other physically or through contribution to mutual processes (e.g. oxidative phosphorylation) so many complex associations exist within proteome data. If key nodes within these networks could be identified objectively this would direct the integration of knowledge to focus attention to pertinent avenues of research. Previously, unsupervised techniques including principal component analysis, hierarchal clustering or self organising maps (k-means clustering) have been used to group together proteins that share similar patterns of expression across biological replicates or experimental cases (e.g. [18]). This approach to finding shared patterns of expression can be useful but the fact that proteins cluster together does not provide evidence that they are components of the same network or that they share dependent relationships. In contrast, conditional independence maps can be used to find networks within complex data based on multivariate associations between nodes (in this case, protein species). Herein we use the CImap package [19] which is a constraint-based approach to discovering multivariate association networks and is used to create a directed acyclic graph of associations within the data. Conceptually, this approach is perfectly matched to the task of interrogating data from techniques such as DIGE, which create highly-detailed parallel information that might reasonably be expected to depict functional, but unknown, associations between protein species.
The current work focuses specifically on skeletal muscle responses to artificial selection on running capacity. Previous studies (e.g. [7]) report the greatest differences in muscle metabolism between HCR and LCR are evident in fast-twitch and mixed-fibre muscles, such as the gastrocnemius. However, the proportion of type IIa/x fibres in gastrocnemius of HCR rats (42 ± 16%) is approximately double (P < 0.01) that (22 ± 14%) of LCR animals [10], and this difference in myofibre profile probably contributes to the differences in muscle metabolism. Here, we investigate whether differences in metabolism between HCR and LCR are evident in the soleus muscle which has a similar myofibre profile in HCR and LCR animals [10]. Of equal importance, the aforementioned literature is based on data from female HCR/LCR animals only. Muscle of females may have a greater capacity for fatty acid metabolism than male muscle [20], therefore, differences in HCR and LCR metabolism might be exaggerated or skewed when studied in an entirely female population. Indeed, just 3 studies [21–23] report skeletal muscle analysis of HCR/LCR males. Moreover, few proteomic studies have addressed the issue of sexual-dimorphism in skeletal muscle. Preliminary data suggest that sex-specific responses may occur in muscle of rats exposed to high-fat diet [24] and streptozotocin administration [25] but these comparisons on pooled samples preclude statistical analysis and interpretation relative to biological variability. Metskas et al. [26] report sex-specific differences in the biceps brachii proteome of mice, including greater abundances of creatine kinase, glycolytic enzymes and components of the pyruvate dehydrogenase complex in males, whereas myoglobin and electron transfer protein alpha were more abundant in females. These findings seem consistent with the reported [27] effects of oestrogen on muscle metabolism but fall short of providing clear evidence of gross differences in mitochondrial density or lipid oxidation. Nonetheless, these limited data raise the issue of whether literature reporting differences in HCR/LCR muscle metabolism are biased by the disproportionate use of female animals, including potential underreporting of paternal traits.
2. Materials and methods
2.1. Animal model
The inception of HCR-LCR strains from a founder population of genetically heterogeneous N:NIH rats has been described in detail [5]. Thirty-two male and female HCR/LCR rats (n = 8, in each group) from generation 25 (12–13 weeks old) were imported from the University of Michigan. The transfer of animals to the UK and subsequent procedures were conducted under the British Home Office Animals (Scientific Procedures) Act 1986 and according to UK Home Office Guidelines. Rats were housed in a conventional facility and the environmental conditions controlled at 20 ± 2 °C, 45–50% relative humidity with a 12 h light (0600–1800) and dark cycle. Food and water were available ab libitum during a 14-day acclimatization period. After an overnight fast, animals were asphyxiated with CO2 and killed by cervical dislocation. Blood was collected by cardiac puncture and allowed to clot at room temperature prior to being placed on ice overnight. After centrifugation, serum fractions were stored at −80 °C and later analysed by ELISA for leptin (Millipore, Billerica, MA). Skeletal muscles and other organs were isolated and cleaned of fat and connective tissue before being weighed. In preparation for histochemical analysis, a segment of the mid-belly of each skeletal muscle was resected and mounted in transverse section before being snap-frozen in supercooled isopentane. Counter lateral muscles were frozen in liquid nitrogen in preparation for proteomic analyses.
2.2. Histochemical analysis of muscle phenotype
Serial cryosections (5 μm thick) were cut from soleus muscle specimens and stained using nicotinamide dinucleotidetetrazolium reductase (NADH-TR) or periodic acid-Schiff (PAS) techniques, described in [28]. Myofibre types were determined based on anti-MyHC type I and IIa (1:10 dilution, N2.261; Axxora) and anti-MyHC type IIa and IIx (1:50 dilution, N3.36; Santa Cruz) Ab staining. Primary Ab was detected with HRP-conjugated secondary Ab (1:100 dilution) and visualised using a DAB and counterstained with haematoxylin. Cryosections were viewed (100 magnification) by light microscopy and were digitised using a 12-bit charge-coupled device (1213C; DVC, Austin, Texas). One hundred myofibres from each muscle were randomly selected and identified as being either type I, type IIa or type IIx/b. Calibrated image analysis software (Lucia; LIM, Hostivar, Czech Republic) was used to measure myofibre cross-sectional area (CSA), and the average mitochondrial density and glycogen content were estimated by measuring the optical density of type I, IIa, or IIx/b fibres (100 each) on NADH-TR or PAS-stained cryosections, respectively.
2.3. DIGE of soluble muscle proteins
Soleus muscles were pulverised in liquid nitrogen then homogenised on ice in 8 volumes of 1% Triton X-100, 50 mM Tris pH 7.4 containing Complete™ protease and PhosSTOP phosphatase inhibitors (Roche Diagnostics, Lewes, UK). Samples were incubated on ice for 10 min then centrifuged at 12,000 rcf, 4 °C for 45 min. Supernates were precipitated in acetone and resuspended in lysis buffer: 7 M urea, 2 M thiourea, 4% (w/v) CHAPS, 30 mM Tris, containing protease and phosphatase inhibitors. Protein concentrations were measured using the Bradford assay (Sigma, Poole, Dorset, UK) and each sample adjusted to 5 μg μl−1 in either Lysis buffer for DIGE analysis or Laemmli buffer for western blot analyses.
Fifty microgram aliquots of each sample and the pooled internal standard were labelled with 400 pM CyDye DIGE Fluor minimal dyes (GE Healthcare, Little Chalfont, UK), consistent with previous work [17]. To minimise the potential confounding effects of differences in fluorescence intensity, Cy3 and Cy5, labelling was alternated between LCR and HCR samples in a ‘balanced’ design. Labelled LCR and HCR aliquots and pooled Cy2-labelled standard were combined with rehydration buffer: 7 M urea, 2 M thiourea, 2% (w/v) CHAPS, 20 mM DTT and 0.5% (v/v) ampholytes. IPG strips (24 cm pH 3– 10 non-linear; Immobiline Drystrip, GE Healthcare) were rehydrated overnight in 450 μl rehydration buffer that contained the combined cyanine-labelled samples. Isoelectric focusing (maximum 50 μA per strip) was performed on an IPGPhor II (GE Healthcare) at 20 °C using the protocol (total 44000 Vh): 3 h at 300 V, 3 h at 600 V, 3 h at 1000 V, gradient to 8000 V in 3 h and then 4 h at 8000 V. IPG strips were equilibrated in 50 mM Tris–HCl pH 8.8, containing 6 M urea, 70 mM SDS, 30% v/v glycerol and a trace amount of bromophenol blue. DTT (65 mM) was present in the first equilibration and iodoacetamide (135 mM) in the second. Proteins were electrophoresed (Ettan Dalt six; GE Healthcare) through denaturing 12.5% poly-acrylamide gels at 20 °C; at 5 W per gel for 30 min, then 17 W per gel until the tracking dye reached the bottom edge.
2.4. Difference profiling and statistical analyses
Gels were digitised (16-bit greyscale, 100 μm pixel size) immediately after electrophoresis using a fluorescence scanner (Typhoon 9400, GE Healthcare) at wavelengths appropriate for Cy2, Cy3 and Cy5 dyes. Gel images (Cy2, Cy3 and Cy5, total = 30) were aligned in SameSpots (Nonlinear Dynamics, Newcastle, UK) using prominent spots (mean ± SD per gel image: 548 ± 36) as vectors to warp each image to a common reference gel. A mask was used to remove gel artefacts (109 spots deleted) and features with an average normalised volume < 10,000 (147 spots) or spot area < 150 (33 spots) were disregarded. In total, 696 spots were included in the subsequent analysis. As a measure of technical variation, when ranked by coefficient of variation, 70% of spots in Cy2 images (pooled standard) had a coefficient of variation less than 14%. Log transformed spot volumes, expressed relative to the pooled standard, were exported to PASW Statistics software (IBM, Portsmouth, UK) and used to investigate differences in expression due to sex (male versus female) and strain (LCR versus HCR) by two-way analysis of variance. To control false discovery rate (FDR), P-value distributions were used to calculate q-values [29] and a criterion FDR of <10% was set. This statistical approach considers the biological variation across each spot and is, therefore, more sophisticated than arbitrarily implementing a threshold based on fold-change.
2.5. Identification of gel spots using nLC-ESI-MS/MS
Proteins were identified from colloidal Coomassie-stained (Bio-Safe; Bio-Rad, Hercules, CA, USA) preparative gels loaded with 1.5 mg protein (pooled standard). A batch of 384 spots was cut and processed using a Xcise robot (Shimadzu Biotech) directed by a picking list produced from the SameSpots analysis. In addition, spots adjacent to differentially expressed proteins were also excised to investigate the presence of different species of these proteins. In-gel tryptic digestion was conducted in 96-well microtitre plates as described previously [4] and the reaction was terminated by the addition of formic acid to a final concentration of 0.1%. Tandem mass spectra were recorded using a quadrupole-high capacity ion-trap (HCT Ultra ETD II; Bruker Daltonics, Bremen, Germany) coupled via an electrospray ionisation source to a nano-flow HPLC system (Ultimate 3000; Dionex, Sunnyvale, CA). Samples (10 μl in-gel digest) were loaded in aqueous 0.1% (v/v) formic acid via a Zorbax 300SB-C18, 5 μm, 5 mm × 300 μm pre-column (Agilent Technology, Santa Carla, CA). Separation was conducted at 30 °C through a Zorbax 300SB-C18, 3.5 μm, 150 mm × 75 μm analytical reverse phase column (Agilent Technology). Peptides were eluted using a gradient rising to 40% acetonitrile 0.1% (v/v) formic acid over 30 min at a flow rate of 300 nl/min. An online nanospray source (EasyNano, Bruker Daltonics) was used equipped with a fused silica emitter. The capillary voltage was −1300 V and a survey scan from 350 m/z to 1600 m/z was used to select peptides with charge states of +2 or +3 using Enhanced Scan mode (8100 (m/z)/s). Data dependent MS/MS analysis in alternating CID and ETD modes was performed selecting the two most abundant precursor ions with active exclusion enabled. Raw data were processed (Data Analysis 4.0, Bruker Daltonics) and Mascot generic format files were searched against the Swiss-Prot database (2011.6) restricted to ‘Rattus’ (7617 sequences) using a locally implemented Mascot (www.matrixscience.com) server (version 2.2.03). A first round Mascot error tolerant search and subsequent directed searches were performed using a bioinformatic platform (ProteinScape, Bruker Daltonics). The enzyme specificity was trypsin allowing 1 missed cleavage, carbamidomethyl modification of cysteine (fixed), oxidation of methionine (variable) and an m/z error of ± 0.5 Da. The directed searches also included acetylation of lysine and phosphorylation of serine, threonine or tyrosine residues as possible modifications.
2.6. Bioinformatic analyses
Functional enrichment analysis was conducted using the Database for Annotation, Visualization and Integrated Discovery [DAVID; http://david.abcc.ncifcrf.gov/home.jsp [30]]. The non-redundant lists of differentially expressed proteins (FDR < 10% for each main effect: sex and strain) were investigated for over-representation of gene ontology (GO) classes: cellular component (CC), biological process (BP) and molecular function (MF). Association of proteins with pathways of the Kyoto Encyclopedia of Genes and Genomes [KEGG; http://www.genome.jp/kegg/ [31]] was also assessed, and the statistical significance of clusters was estimated by modified Fisher exact P value.
Conditional independence mapping was used to graph interrelations within the population of spots that were significantly (FDR < 10%) differentially expressed due to HCR/LCR strain. Data were analysed in MatLab using the CImap package [19] to create conditional independence maps that describe multivariate association networks within the data. Log-transformed continuous data of spot expression were converted to categorical terciles or quintiles to assess course- and fine-grain associations, respectively. Each analysis began with a fully-connected undirected graph and conditional independence tests were used to determine whether edges (associations) between vertices (protein spots) should be kept or deleted. These decisions were made using conditional mutual information, which is an estimate of association strength that is approximated by the chi-squared distribution. That is, decisions regarding inclusion or exclusion of edges were made by performing chi-squared tests on the null hypothesis that two vertices are independent. This process was ordered so as to test and exclude weak edges first and arrive at a directed acyclic graph that depicts the multivariate association network within the data. CImap also includes computational procedures for bounding the false positive and false negative rates, to enable conditional independence testing within small sample sizes. To construct the tercile map α was set at 0.05, whereas more stringent (α = 0.01) testing was used in the construction of the quintile map. Post-hoc pair-wise testing was used to approximate the relative strength of associations between vertices in order to dictate edge length (shorter edge = stronger association) during the construction of each map.
Bibliometric network mapping was conducted using iHOP (http://www.ihop-net.org; [32]). Gene models were constructed from evidence in the rat, human and mouse interaction pages for the protein of interest. Information was manually curated and filtered to include evidence of co-expression, co-localisation, protein–protein interaction and modulation by gain/loss of function interventions.
2.7. Western blotting
Samples in Laemmeli buffer [33] were heat denatured (5 min at 95 °C) and aliquots containing 50 μg protein were loaded on precast 4–20% polyacrylamide gels (Precise Tris–HEPES protein gels; Thermo Scientific). Electrophoresis was conducted at 80 V for 20 min then 140 V for 40 min using a mini-gel system (MiniVE; GE Healthcare, Little Chalfont, UK). Electro-transfer of proteins to methanol-activated polyvinylidene difluoride membranes was conducted in Towbin buffer [34] at 25 V for 2 h. Successful transfer was confirmed by staining membranes with direct blue 71, according to [35]. Non-specific protein interactions were blocked by incubating the membranes with 5% non-fat dry milk in 20 mM Tris, 150 mM NaCl, and 0.1% Tween 20, pH 7.6 (TBS-T) for 1 h at room temperature. Membranes were then washed in TBS-T and incubated overnight with TBS-T containing 5% BSA and primary antibodies specific for: signal transducer and activator of transcription 3 (STAT3; 9139 Cell Signalling Technology; 1:5,000 dilution) and phosphorylated (S727 or Y705) STAT3 (9134 and 9131, Cell Signalling Technology; 1:500 dilution). Serial washes in TBS-T were performed prior to enhanced chemiluminescence (ECL Prime; GE Healthcare) and digitization (Gel Doc XRS; Bio-Rad, Hercules, CA) of immuno-reactive protein bands. Image analysis (Quantity One, version 4.; Bio-Rad) was used to measure the relative abundances of target proteins. Analysis of phosphorylated and non-phosphorylated species was achieved by stripping and re-probing of membranes. Briefly, membranes were stripped by incubation in 62.5 mM Tris, 70 mM SDS, 50 mM β-mercaptoethanol, pH 6.8 at 50 °C for 30 min. Successful removal of the immuno-signal was assessed by enhanced chemiluminescence prior to subsequent immuno-detection.
3. Results
3.1. Physical and physiological characteristics
The physical characteristics of male and female HCR and LCR rats are displayed in Table 1. In females, body weight of LCR was 1.26-fold greater than HCR, and the body weight of males was 1.38-fold greater in LCR compared to HCR. There was a statistically significant interaction in distance completed during a standardised exercise test. HCR females ran significantly further than any other group and the difference in running capacity between HCR and LCR strains was 5.87-fold in females and 6.44-fold in males. Hence, regardless of strain, females were lighter and ran further than males and this corresponded with significant differences in work (J) performed during the exercise test. Consistent with the smaller body mass of females the wet weight of the heart and skeletal muscles was significantly less in females compared to males. Similarly, absolute wet weights of heart and red gastrocnemius muscle were significantly less in HCR (i.e. consistent with the differences in body weight), but there was no significant difference in the average wet weights of soleus, EDL and tibialis anterior muscle between HCR and LCR strains.
Table 1.
Physical characteristics of male and female LCR and HCR rats.
| F-LCR | M-LCR | F-HCR | M-HCR | Interaction | Strain | Sex | |
|---|---|---|---|---|---|---|---|
| Distance run (m) | 336.5 ± 52.9 | 252.3 ± 42.7 | 1974.1 ± 108.6 | 1624.5 ± 112.1 | 0.0001 | <0.0001 | <0.0001 |
| Work (J) | 176.7 ± 30.9 | 219.8 ± 43.9 | 853.1 ± 53.9 | 1019.6 ± 93.7 | 0.0074 | <0.0001 | <0.0001 |
| Body weight (g) | 255.4 ± 27.2 | 460.4 ± 40.4 | 203.0 ± 16.7 | 334.4 ± 35.8 | 0.0025 | <0.0001 | <0.0001 |
| Soleus weight (mg) | 111.4 ± 12.8 | 157.3 ± 32.5 | 116.3 ± 10.1 | 162.5 ± 23.3 | NS | NS | <0.0001 |
| EDL weight (mg) | 109.5 ± 9.2 | 176.1 ± 17.4 | 107.3 ± 17.6 | 171.3 ± 19.9 | NS | NS | <0.0001 |
| Red gastrocnemius weight (mg) | 521.1 ± 56.2 | 732.9 ± 96.6 | 480.3 ± 80.6 | 600.4 ± 164.2 | NS | 0.0299 | <0.0001 |
| Tibialis anterior weight (mg) | 452.1 ± 50.3 | 693.3 ± 86.9 | 478.0 ± 79.9 | 633.3 ± 105.6 | NS | NS | <0.0001 |
| Heart weight (mg) | 789.4 ± 76.4 | 1229.6 ± 119.7 | 707.9 ± 44.3 | 1090.3 ± 100.1 | NS | 0.0016 | <0.0001 |
Data are displayed as mean ± SD (n = 5, in each group). Two-factor analysis of variance was used to calculate P values and determine statistically significant interactions and main effects for strain (HCR vs LCR) and sex (male vs female). EDL = extensor digitorum longus, F-HCR = female high-capacity runner, M-LCR = male low-capacity runner.
3.2. Soleus muscle phenotype
Soleus had type I (~20%) and type IIa (~80%) myofibres only and there was no significant difference in the portions of these fibres between male and female HCR and LCR. The CSA of type I and IIa fibres was greater in male than female, this was statistically significant for type IIa fibres and consistent with the greater soleus mass in males compared to females. The area fractions of type I and type IIa were not different in male and female HCR and LCR soleus (Table 2). Mitochondrial density (NADH-TR staining) of both type I and type IIa fibres was significantly greater in HCR than LCR. No significant differences were detected in myofibre glycogen content (PAS staining).
Table 2.
Soleus phenotype of male and female LCR and HCR rats.
| F-LCR | M-LCR | F-HCR | M-HCR | Interaction | Strain | Sex | |
|---|---|---|---|---|---|---|---|
| Proportion of type I fibres (%) | 84 ± 11 | 78 ± 7 | 75 ± 15 | 80 ± 12 | NS | NS | NS |
| Cross sectional area (mm2) | |||||||
| Type I | 2560 ± 290 | 3037 ± 627 | 2674 ± 223 | 3004 ± 510 | NS | NS | 0.0589 |
| Type IIa | 1733 ± 969 | 3318 ± 711 | 1795 ± 367 | 2590 ± 572 | NS | NS | 0.0014 |
| NADH-TETRAZOLIUM reductase (OD) | |||||||
| Type I | 0.158 ± 0.03 | 0.146 ± 0.03 | 0.169 ± 0.01 | 0.192 ± 0.01 | NS | 0.0244 | NS |
| Type IIa | 0.161 ± 0.04 | 0.148 ± 0.04 | 0.187 ± 0.02 | 0.207 ± 0.01 | NS | 0.0059 | NS |
| Periodic acid Schiff's (OD) | |||||||
| Type I | 0.183 ± 0.07 | 0.179 ± 0.02 | 0.177 ± 0.02 | 0.193 ± 0.01 | NS | NS | NS |
| Type IIa | 0.187 ± 0.02 | 0.186 ± 0.08 | 0.172 ± 0.03 | 0.197 ± 0.01 | NS | NS | NS |
Data are displayed as mean ± SD (n = 5, in each group). Two-factor analysis of variance was used to calculate P values and determine statistically significant interactions and main effects for strain (HCR vs LCR) and sex (male vs female). EDL = extensor digitorum longus, F-HCR = female high-capacity runner, M-LCR = male low-capacity runner.
3.3. Proteome mining of soluble skeletal muscle proteins
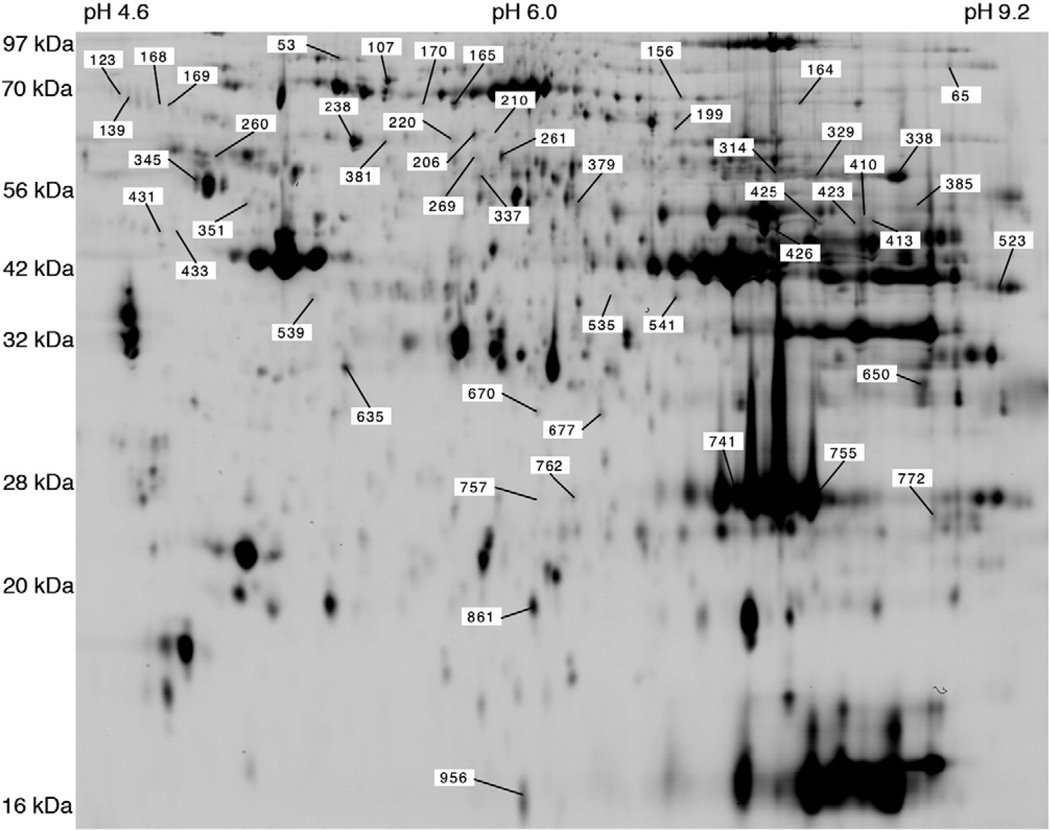
Mass spectrometry of selected 2D gel spots unambiguously identified proteins in 337 spots and generated a non-redundant list of 154 gene products. A detailed 2D gel map of the proteins identified by mass spectrometry is available in the World 2D-PAGE repository (http://world-2dpage.expasy.org/repository/), accession number 0069. This resource displays the position and Swiss-Prot database identification of each gel spot. The spot reference numbers used in this article are also displayed and each identified spot is linked to the raw CID and ETD MS/MS spectra in Mascot Generic Format (.mgf).
Based on the outcomes of Mascot error tolerant searches, directed MS/MS ion searches were performed with phosphorylation (S/T and Y) and acetylation (K) as possible modifications. Manual verification of fragment-ion sequence ladders in peptides with scores >20 created a list of 70 peptides from 29 gel spots, which encompassed 19 non-redundant gene products (Supplementary Table S1). Of the 70 modified peptides, 59 have been previously reported and 11 were novel modifications not currently recorded in PhosphoSite (www.phosphosite.org). In the majority, potential sites of modification were supported by CID spectra only, which may be a consequence of the relatively short LC gradient, arbitrary ordering of CID before ETD fragmentation or the preference of ETD fragmentation for higher charge-state precursors. Five spots (411, 419, 427, 431, 433) each identified as N-myc down-regulated gene 2 (NDRG2) protein, exhibited CID evidence of numerous S/T phosphorylation sites. In each of the NDRG2 spots, CID and ETD fragment spectrum pairs for a peptide spanning residues 328–343 and indicating phosphorylation of serine 228, 230 or 332 were recorded during the proteome mining exercise.
3.4. DIGE profiling of male and female, HCR and LCR soleus
Two-way analysis of variance performed on the 696 gel spots matched in male and female soleus muscle of HCR and LCR rats detected 15 spots that were significantly (P < 0.05, FDR < 10%) different between males and females (Table 3) and 40 spots that were significantly (P < 0.05, FDR < 10%) different between HCR and LCR strains (Table 4). An image of the 2D reference gel annotated with the position of each of the statistically different protein spots is shown in Fig. 1. Overlap of spots that were different between HCR/LCR strain and those different between sexes was limited to 4 spots (#53 HSP7C, #139 SPA3K, #165 ODP2 and #379 ENOB) and just one spot (#379, β-enolase) exhibited a statistically significant interaction (more abundant in male LCR). β-Enolase is a highly abundant glycolytic enzyme and was resolved into 9 spots that had different pI but similar Mr. This spot pattern may reflect different states of modification. β-Enolase can undergo S, T and Y phosphorylation and K acetylation but proteome mining did not identify post-translational modifications specific to spot #379. Moreover, the 1.06-fold greater abundance of spot #379 specific to male LCR was somewhat overshadowed by the 2.57-fold greater abundance of β-enolase spot #410 that was observed in both male and female LCR (Table 4). Considering the wider effects on metabolic enzymes (Fig. 2), the greater abundance of β-enolase likely reflects a shift toward greater reliance on glycolytic metabolism in LCR muscle.
Table 3.
Sex-specific differences in soleus muscle protein abundance.
| Description | Protein ID | Ref# | F-LCR | M-LCR | F-HCR | M-HCR | Fold diff. |
|---|---|---|---|---|---|---|---|
| Spots significantly more abundant in males | |||||||
| 60 kDa heat shock protein, mitochondrial | CH60 | 381 | 1.414 ± 0.094 | 0.746 ± 0.116 | 1.440 ± 0.271 | 0.755 ± 0.120 | 1.71 |
| Glutathione S-transferase | GSTA3 | 772 | 1.174 ± 0.156 | 0.918 ± 0.209 | 1.262 ± 0.243 | 0.871 ± 0.192 | 1.44 |
| aDesmin | DESM | 351 | 1.070 ± 0.169 | 0.896 ± 0.095 | 1.174 ± 0.133 | 0.963 ± 0.107 | 1.28 |
| Carbonic anhydrase 3 | CAH3 | 741 | 0.959 ± 0.089 | 0.803 ± 0.117 | 0.994 ± 0.147 | 0.772 ± 0.110 | 1.24 |
| Carbonic anhydrase 3 | CAH3 | 755 | 0.865 ± 0.067 | 0.749 ± 0.092 | 0.865 ± 0.113 | 0.691 ± 0.110 | 1.15 |
| Superoxide dismutase [Cu–Zn] | SODC | 956 | 1.173 ± 0.067 | 1.064 ± 0.064 | 1.106 ± 0.078 | 0.971 ± 0.072 | 1.13 |
| aPhosphoglucomutase-1 | PGM1 | 199 | 1.031 ± 0.052 | 0.807 ± 0.102 | 0.930 ± 0.043 | 0.873 ± 0.049 | 1.10 |
| Spots significantly more abundant in females | |||||||
| Serine protease inhibitor A3K | SPA3K | 139 | 1.098 ± 0.278 | 1.351 ± 0.141 | 0.735 ± 0.057 | 1.031 ± 0.112 | 1.33 |
| Serine protease inhibitor A3L | SPA3L | 168 | 0.958 ± 0.278 | 1.220 ± 0.210 | 0.747 ± 0.144 | 1.282 ± 0.125 | 1.28 |
| aAlpha-2-HS-glycoprotein | FETUA | 169 | 0.978 ± 0.171 | 1.306 ± 0.166 | 0.882 ± 0.192 | 1.308 ± 0.184 | 1.29 |
| Heat shock cognate 71 kDa protein | HSP7C | 53 | 0.932 ± 0.105 | 1.167 ± 0.160 | 1.155 ± 0.096 | 1.329 ± 0.143 | 1.23 |
| Stress-70 protein, mitochondrial | GRP75 | 170 | 0.815 ± 0.068 | 1.006 ± 0.115 | 0.935 ± 0.069 | 1.038 ± 0.099 | 1.17 |
| Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex |
ODP2 | 165 | 0.970 ± 0.043 | 1.138 ± 0.090 | 1.113 ± 0.034 | 1.222 ± 0.065 | 1.12 |
| Beta-enolase | ENOB | 379 | 0.879 ± 0.063 | 1.084 ± 0.104 | 0.870 ± 0.020 | 0.858 ± 0.018 | 1.07 |
| Vimentin | VIME | 260 | 0.941 ± 0.215 | 1.238 ± 0.209 | 0.903 ± 0.140 | 1.209 ± 0.121 | 1.13 |
Protein description and protein ID relate to the Swiss-Prot database entry identified from Mascot searches of CID and ETD MS/MS spectra. Ref# is the gel spot number created during image analysis and is consistent with Fig. 1 and the World-2DPAGE online graphical interface, available at http://world-2dpage.expasy.org/repository/0069/. Relative spot volumes (mean ± SD) are reported for each independent group, i.e. male high-capacity runner (M-HCR), female low-capacity runner (F-LCR) etc. Fold difference values are reported for spots exhibiting significant differences in expression at P < 0.05 at a false discovery rate of <10%.
Contains evidence of post-translational modification: acetylation or phosphorylation (Supplementary Table S1).
Table 4.
HCR/LCR strain-specific differences in soleus muscle protein abundance.
| Description | Protein ID | Ref# | F-LCR | M-LCR | F-HCR | M-HCR | fold diff. |
|---|---|---|---|---|---|---|---|
| Respiratory chain | |||||||
| NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 10, mitochondrial |
NDUAA | 535 | 0.643 ± 0.292 | 0.721 ± 0.229 | 1.204 ± 0.302 | 1.182 ± 0.045 | −2.06 |
| ATP synthase subunit alpha | ATPA | 338 | 0.956 ± 0.044 | 1.007 ± 0.098 | 1.100 ± 0.062 | 1.132 ± 0.100 | −1.08 |
| ATP synthase subunit alpha | ATPA | 314 | 0.936 ± 0.048 | 0.967 ± 0.080 | 1.154 ± 0.094 | 1.180 ± 0.085 | −1.18 |
| aATP synthase subunit beta | ATPB | 345 | 0.861 ± 0.028 | 0.929 ± 0.059 | 1.043 ± 0.059 | 1.012 ± 0.113 | −1.10 |
| ATP synthase subunit d | ATP5H | 861 | 1.054 ± 0.052 | 1.134 ± 0.122 | 1.191 ± 0.080 | 1.289 ± 0.107 | −1.04 |
| Mitochondrial protein import | |||||||
| a60 kDa heat shock protein, mitochondrial | CH60 | 238 | 1.109 ± 0.112 | 1.047 ± 0.071 | 1.258 ± 0.117 | 1.170 ± 0.044 | −1.10 |
| aStress-70 protein, mitochondrial | GRP75 | 107 | 1.160 ± 0.078 | 1.061 ± 0.027 | 1.237 ± 0.070 | 1.190 ± 0.077 | −1.08 |
| aHeat shock cognate 71 kDa protein | HSP7C | 53 | 0.932 ± 0.105 | 1.167 ± 0.160 | 1.155 ± 0.096 | 1.329 ± 0.143 | −1.11 |
| Pyruvate dehydrogenase complex | |||||||
| Pyruvate dehydrogenase E1 component subunit beta | ODPB | 635 | 0.991 ± 0.043 | 1.018 ± 0.055 | 1.132 ± 0.026 | 1.181 ± 0.099 | −1.10 |
| Dihydrolipoyllysine-residue acetyltransferase E2 component of pyruvate dehydrogenase complex |
ODP2 | 165 | 0.970 ± 0.043 | 1.138 ± 0.090 | 1.113 ± 0.034 | 1.222 ± 0.065 | −1.08 |
| Tricarboxylic acid cycle | |||||||
| Fumarate hydratase | FUMH | 423 | 0.599 ± 0.304 | 0.508 ± 0.198 | 1.458 ± 0.254 | 1.622 ± 0.335 | −3.12 |
| Fumarate hydratase | FUMH | 425 | 0.768 ± 0.121 | 0.822 ± 0.120 | 1.325 ± 0.177 | 1.374 ± 0.0115 | −1.54 |
| Fumarate hydratase | FUMH | 426 | 0.717 ± 0.269 | 0.599 ± 0.163 | 1.471 ± 0.282 | 1.595 ± 0.222 | −2.35 |
| Fatty acid metabolism | |||||||
| Very long-chain specific acyl-CoA dehydrogenase | ACADV | 65 | 0.878 ± 0.173 | 0.866 ± 0.121 | 1.074 ± 0.049 | 1.074 ± 0.071 | −1.32 |
| Enoyl-CoA hydratase | ECHM | 762 | 0.890 ± 0.064 | 0.885 ± 0.078 | 1.158 ± 0.100 | 1.026 ± 0.074 | −1.20 |
| Amino acid metabolism | |||||||
| 3-mercaptopyruvate sulfurtransferase | THTM | 670 | 0.899 ± 0.040 | 0.883 ± 0.047 | 1.1074 ± 0.079 | 0.969 ± 0.043 | −1.18 |
| b3-hydroxyisobutyrate dehydrogenase | 3HIDH | 677 | 1.031 ± 0.065 | 1.025 ± 0.044 | 1.185 ± 0.030 | 1.154 ± 0.134 | −1.13 |
| abAspartate aminotransferase, mitochondrial | AATM | 523 | 0.809 ± 0.061 | 0.848 ± 0.072 | 1.039 ± 0.170 | 0.922 ± 0.053 | −1.16 |
| Ketone body metabolism | |||||||
| Succinyl-CoA:3-ketoacid-coenzyme A transferase 1 | SCOT1 | 164 | 0.930 ± 0.104 | 0.947 ± 0.093 | 1.093 ± 0.051 | 1.082 ± 0.084 | −1.17 |
| Glycolysis | |||||||
| Beta-enolase | ENOB | 341 | 1.104 ± 0.033 | 0.996 ± 0.053 | 1.146 ± 0.077 | 1.134 ± 0.099 | −1.07 |
| Beta-enolase | ENOB | 410 | 2.121 ± 1.562 | 1.569 ± 0.435 | 1.017 ± 0.246 | 0.530 ± 0.247 | +2.57 |
| Beta-enolase | ENOB | 379 | 0.879 ± 0.063 | 1.084 ± 0.104 | 0.870 ± 0.020 | 0.858 ± 0.018 | +1.06 |
| Glyceraldehyde-3-phosphate dehydrogenase | G3P | 413 | 1.553 ± 0.374 | 1.538 ± 0.409 | 0.679 ± 0.457 | 0.426 ± 0.082 | +2.76 |
| Cytoskeletal | |||||||
| Tripartite motif-containing protein 72 | TRI72 | 261 | 1.272 ± 0.112 | 1.143 ± 0.116 | 0.851 ± 0.229 | 0.869 ± 0.127 | +1.28 |
| Tripartite motif-containing protein 72 | TRI72 | 269 | 0.655 ± 0.065 | 0.689 ± 0.091 | 1.306 ± 0.700 | 1.2302 ± 0.338 | −1.93 |
| Lamin-A | LMNA | 329 | 0.979 ± 0.033 | 1.015 ± 0.061 | 1.115 ± 0.064 | 1.139 ± 0.117 | −1.07 |
| Miscellaneous | |||||||
| Protein disulphide-isomerase A3 | PDIA3 | 206 | 1.808 ± 0.506 | 1.765 ± 1.051 | 0.424 ± 0.106 | 0.448 ± 0.070 | +2.83 |
| Protein disulphide-isomerase A3 | PDIA3 | 220 | 1.727 ± 0.426 | 1.464 ± 0.635 | 1.084 ± 0.146 | 0.722 ± 0.125 | +1.57 |
| bProtein disulphide-isomerase A3 | PDIA3 | 210 | 0.774 ± 0.399 | 0.669 ± 0.265 | 1.139 ± 0.150 | 1.146 ± 0.118 | −1.75 |
| abProtein NDRG2 | NDRG2 | 433 | 0.872 ± 0.191 | 0.985 ± 0.072 | 1.189 ± 0.186 | 1.222 ± 0.152 | −1.28 |
| abProtein NDRG2 | NDRG2 | 431 | 0.890 ± 0.197 | 0.920 ± 0.102 | 1.165 ± 0.092 | 1.098 ± 0.127 | −1.37 |
| Serine protease inhibitor A3K | SPA3K | 123 | 1.159 ± 0.316 | 1.435 ± 0.261 | 0.777 ± 0.130 | 1.008 ± 0.246 | +1.53 |
| Serine protease inhibitor A3K | SPA3K | 139 | 1.098 ± 0.278 | 1.351 ± 0.141 | 0.735 ± 0.057 | 1.031 ± 0.112 | +1.43 |
| Voltage-dependent anion-selective channel protein 1 | VDAC1 | 385 | 1.317 ± 0.320 | 1.245 ± 0.249 | 0.944 ± 0.128 | 0.830 ± 0.191 | +1.33 |
| Voltage-dependent anion-selective channel protein 1 | VDAC1 | 650 | 0.945 ± 0.086 | 0.973 ± 0.090 | 1.102 ± 0.119 | 1.166 ± 0.035 | −1.16 |
| Aspartyl-tRNA synthetase, cytoplasmic | SYDC | 156 | 0.831 ± 0.103 | 0.928 ± 0.079 | 1.022 ± 0.059 | 1.084 ± 0.111 | −1.19 |
| Aldehyde dehydrogenase, mitochondrial | ALDH2 | 337 | 1.058 ± 0.079 | 1.013 ± 0.102 | 1.176 ± 0.100 | 1.253 ± 0.063 | −1.16 |
| Creatine kinase M-type | KCRM | 541 | 0.738 ± 0.242 | 0.853 ± 0.238 | 1.275 ± 0.227 | 1.303 ± 0.279 | −1.79 |
| Hypoxanthine-guanine phosphoribosyltransferase | HPRT | 757 | 0.768 ± 0.138 | 0.822 ± 0.052 | 1.060 ± 0.082 | 0.874 ± 0.114 | −1.31 |
| Serine protease inhibitor A3L | SPA3L | 539 | 1.557 ± 0.248 | 1.688 ± 0.395 | 0.762 ± 0.657 | 0.311 ± 0.055 | +3.18 |
Protein description and protein ID relate to the Swiss-Prot database entry identified from Mascot searches of CID and ETD MS/MS spectra. Ref# is the gel spot number created during image analysis and is consistent with Fig. 1 and the World-2DPAGE online graphical interface, available at http://world-2dpage.expasy.org/repository/0069/. Relative spot volumes (mean ± SD) are reported for each independent group, i.e. male high-capacity runner (M-HCR), female low-capacity runner (F-LCR) etc. Fold difference values are reported for spots exhibiting significant differences in expression at P < 0.05 at a false discovery rate of <10%.
Present in Table S3 from {Bell 2009} containing genes from the longevity interactome (core or primary interactors) that are differentially expressed in young versus old human muscle.
Contains evidence of post-translational modification: acetylation or phosphorylation (Supplementary Table S1).
Fig. 1.
DIGE reference map. Reference 2D gel map of rat soleus muscle. Annotations show the reference numbers of spots that were significantly different between either male vs females (Table 3) or high-capacity runners (HCR) vs low-capacity runners (LCR; Table 4). An interactive version of the gel map is available at the World-2DPAGE repository, accession number 0069.
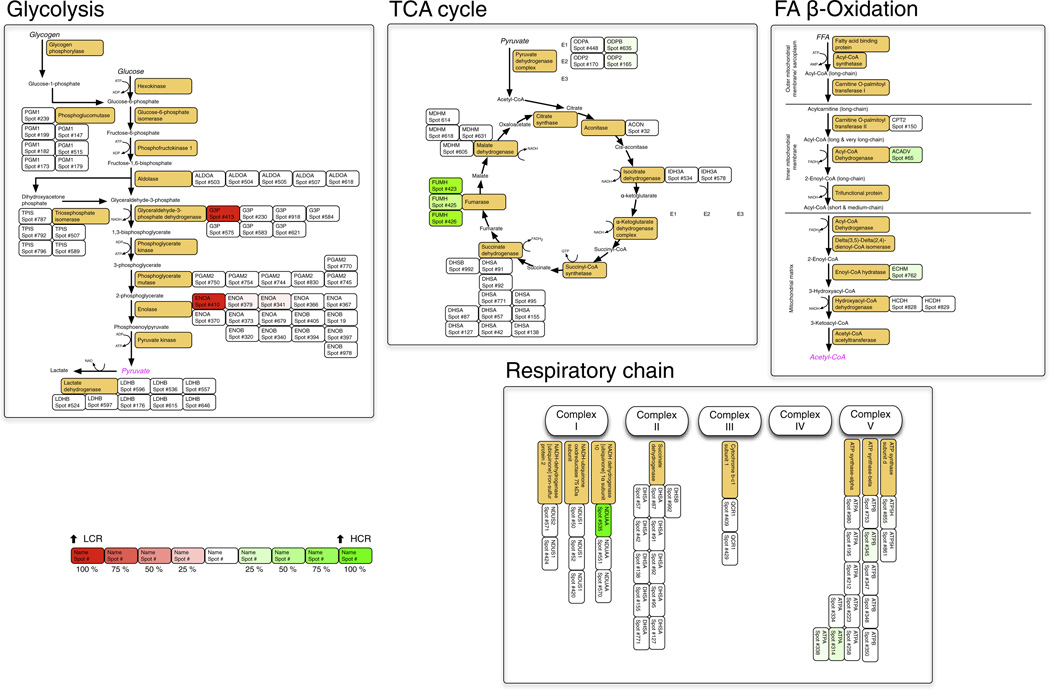
Fig. 2.
Metabolic maps. The metabolic pathways of glycolysis, fatty acid β-oxidation and the tricarboxylic acid cycle are redrawn from the Kyoto Encyclopedia of Genes and Genomes (KEGG). For clarity the respiratory chain is not shown in its entirety, instead only subunits detected by DIGE are highlighted. Orange boxes display the common name of each enzyme, the adjacent boxes detail protein spots matched to this UniProt Rattus ID by LC–MS/MS analysis. A ‘heat map’ colouring system is used to display proteins that were significantly more abundant in HCR (green) or LCR (red) groups (Table 4).
Functional annotation linked the non-redundant protein identifications with several GO classes and KEGG pathways. Proteins more abundant in HCR (total 25 non-redundant gene products) linked with mitochondria (CC), generation of precursor metabolites (BP), nucleotide binding (MF) and were associated with TCA Cycle and Ox Phos KEGG pathways (Fig. 2). Proteins more abundant in LCR (total 7 non-redundant gene products) linked with serine-type endopeptidase inhibitor activity (MF) and were associated with the Glycolysis KEGG pathway. The main sex-specific difference was the enrichment of cytosolic proteins (CC) linked with the response to oxidative stress (BP) and chaperone binding (MF) in males (total 8 non-redundant gene products). The 7 non-redundant gene products that were more abundant in females did not exhibit significant enrichment of GO or KEGG classifications.
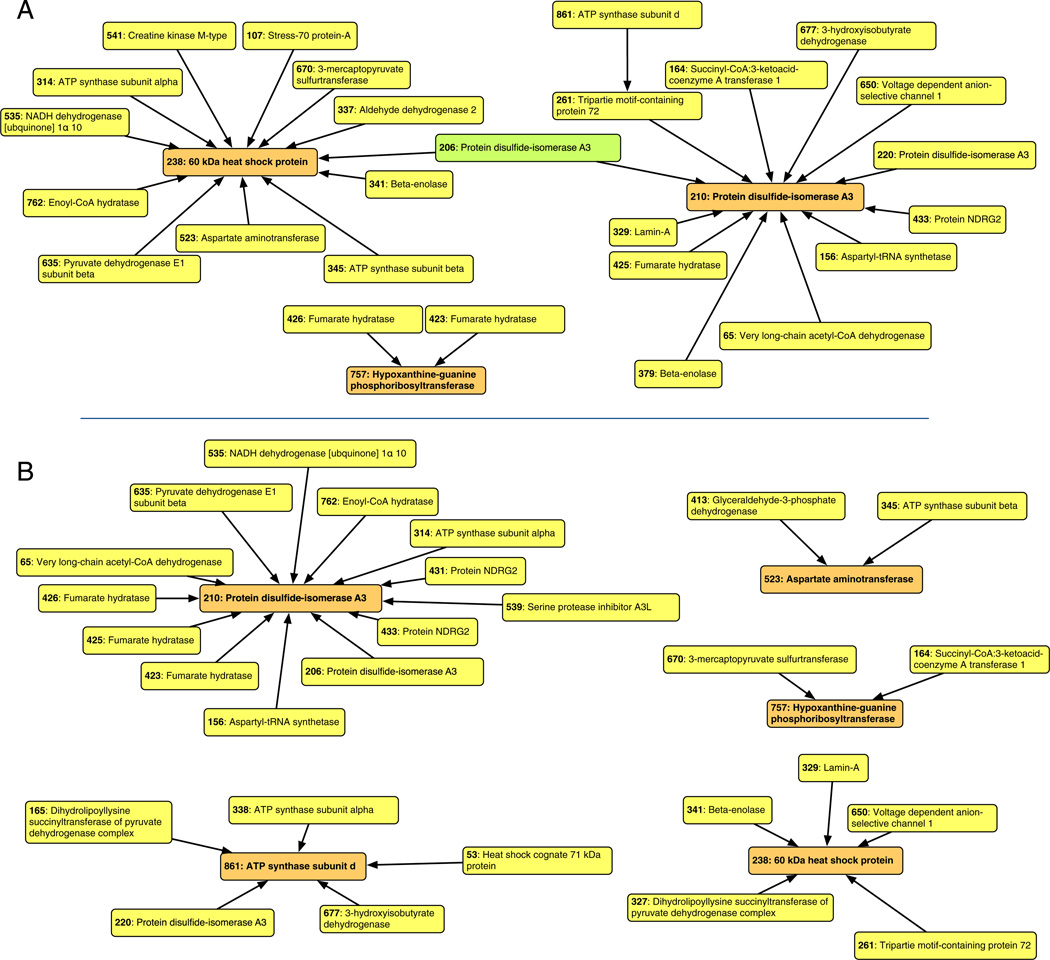
Conditional independence maps of course- and fine-grain associations within the DIGE data were complementary with the functional enrichment analysis and also highlighted important additional information. Course-grain mapping (Fig. 3A) based on significant (α = 0.05) associations found using tercile categorisation of the data highlighted 3 clusters focusing on 60 kDa heat shock protein (CH60; spot #328), protein disulphide isomerase A3 (PDIA3; spot #210) and hypoxanthine–guanine phosphoribosyltransferase (HPRT; spot #757). The cluster centred on CH60 primarily consisted of mitochondrial proteins, which is consistent with the functional enrichment analysis and the key role of CH60 in mitochondrial protein import. A second PDIA3 spot (#206) linked the two principal clusters of CH60/spot #238 and PDIA3/spot #210. Thus protein species of PDIA3 were connected with a large number of the differentially regulated proteins and may also provide a link to the different mitochondrial and aerobic capacities of HCR and LCR muscle. PDIA3 (spot #210) became the most prominent feature when more stringent (α = 0.01) conditional independence mapping was performed on quintile categorised data (Fig. 3B) but the link with theCH60was broken. The PDIA3 cluster included, amongst others, PDIA3 spot (#206) and serine protease inhibitor A3K (spot #539), which was the spot exhibiting the greatest difference (3.18-fold) between HCR and LCR muscle. Post-hoc testing revealed that the strongest associations with PDIA3 spot #210 wereNDRG2 (spot #433; K = 1.1761), fumarate hydratase (FUMH, spot #426; K = 1.1761) and very long-chain acetyl CoA dehydrogenase (ACADV, spot #65; K = 1.1573).
Fig. 3.
Conditional independence maps of tercile and quintile associations. Conditional independence mapping was used to find multivariate association networks within the protein spots differentially expressed between HCR and LCR muscle (Table 4). Log-transformed continuous data of spot expression were converted to categorical terciles (A) or quintiles (B) to assess course- and fine-grain associations, respectively. To construct the tercile map α was set at 0.05, whereas more stringent (α = 0.01) testing was used in the construction of the quintile map. Post-hoc pair-wise testing was used to approximate the relative strength of associations between vertices in order to dictate edge length (shorter edge = stronger association) during the construction of each map. Spot numbers and protein names correspond with Fig. 1, Table 4 and the World-2DPAGE database (accession #0069).
Proteome mining showed that PDIA3 was resolved as a total of 4 gel spots, and 3 of the protein species were differentially expressed between HCR and LCR (Table 4). Spot #206 and #220 were more abundant in LCR whereas spot #210 was greater in HCR, which indicate a shift in post-translational state. Routine mass spectrometry analysis conducted during the proteome mining exercise identified potential phosphorylation of S176 in PDIA3 spot #210 (see Supplementary data).
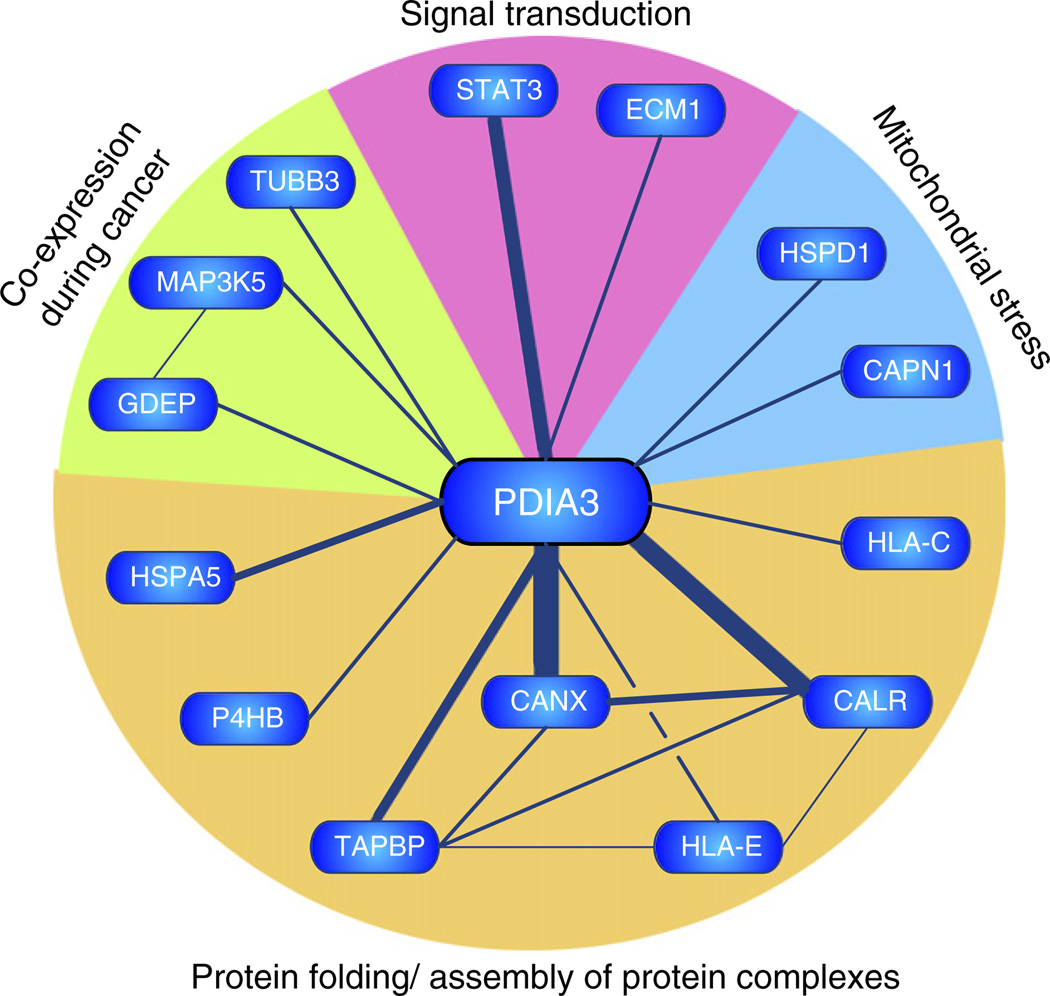
A bibliometric network was constructed as an unbiased means to explore literature evidence of proteins that have been associated with PDIA3 (Fig. 4). The majority of citations were related to protein–protein interactions between PDIA3 and the ER chaperones calnexin and calreticulin. Second to these interactions was strong evidence of the association of PDIA3 with the signal transducer and activator of transcription 3 (STAT3), which encompassed evidence from co-expression, co-localisation, protein–protein interaction and gain/loss of function studies.
Fig. 4.
Bibliometric network of PDIA3. A gene model representing the bibliometric network of PDIA3 was constructed by automated text-mining (data redrawn from iHOP; information hyperlinked over proteins; www.ihop-net. org). Nodes (genes) are connected by edges, which represent co-occurrence within sentences of peer-reviewed published literature. Edge thickness approximates the number of supporting sentences. The majority of associations (CANX, calnexin; CALR, calreticulin, TAPBP, tapsin; HLA-E/HLA-C, major histocompatibility complex IE/C; HSPA5, glucose regulated protein 78; P4HB, prolyl-4-hydroxylase beta) were related to protein folding or assembly of protein complexes within the endoplasmic reticulum. PDIA3 was also reported to interact (TUBB3, class III beta tubulin) or be co-expressed with mitogen activated protein kinase kinase kinase 5 (MAP3K5) and gene differentially expressed in prostate (GDEP) in malignant stages of prostate cancer. Fourteen sentences reported interaction or colocalisation of PDIA3 with STAT3.
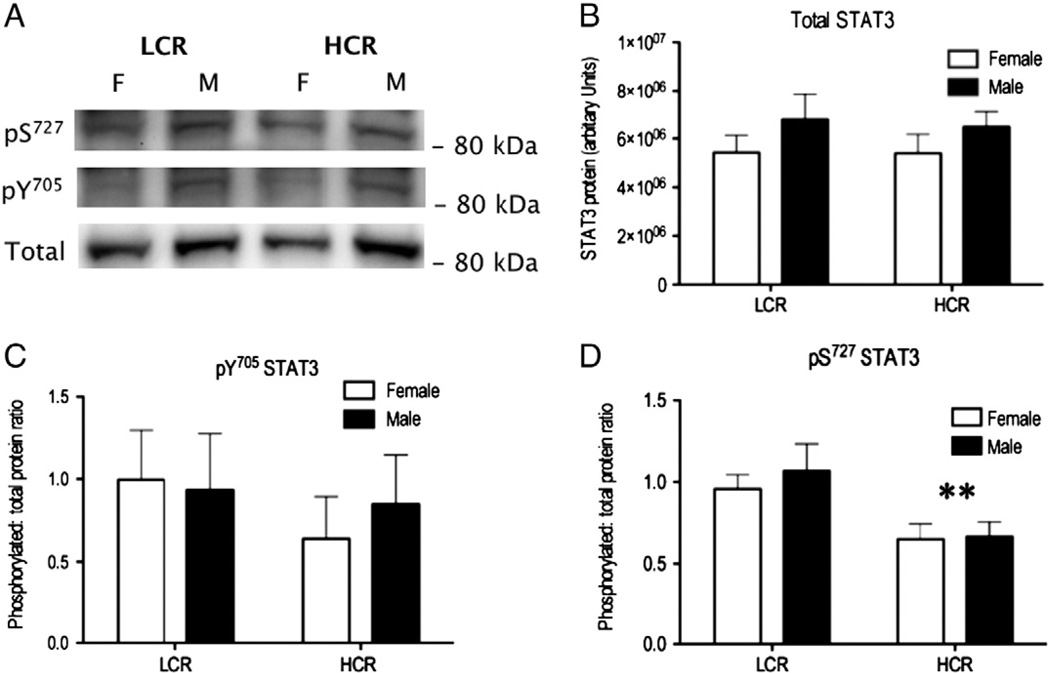
3.5. Western blot analysis of STAT3
The abundance of STAT3 protein was not significantly different between male and female or HCR and LCR soleus muscles (Fig. 5). Similarly, there was no significant difference in the relative ratio of tyrosine (Y705) phosphorylation of STAT3. However, serine (S727) phosphorylation of STAT3 was 1.54-fold greater (P = 0.0058) in LCR soleus.
Fig. 5.
STAT3 phosphorylation in male and female HCR/LCR muscle. STAT3 protein abundance and relative phosphorylation of either tyrosine (Y705) or serine (S727) residues was measured in the solei of male and female, HCR and LCR animals (A). Two-way analysis of variance found no significant difference in the abundance of STAT3 (B) or in the relative amount of Y705 phosphorylation (C). In contrast, a significant (*P < 0.05) main effect of ‘strain’ (i.e. HCR vs LCR) was evident for S727 phosphorylation. On average, the extent of S727 phosphorylation was 1.54-fold greater in LCR soleus (D).
3.6. Serum leptin levels
STAT3 is a canonical component of skeletal muscle leptin signalling and differences in serum leptin levels in HCR/LCR animals have been reported [36]. There were significant (P < 0.05) sex-specific and strain-specific differences in serum leptin concentrations. Values in males (M-HCR = 2.92 ± 1.56 ng·ml−1; M-LCR = 5.71 ± 2.59 ng·ml−1) were greater than values in females (F-HCR = 1.07 ± 0.46 ng·ml−1; F-LCR = 2.76 ± 0.71 ng·ml−1) but there was no statistically significant interaction between sex and strain. Further to differences between HCR and LCR group means, significant correlation was found between individual serum leptin concentrations in LCR and the abundance of 60 kDa heat shock protein (spot #238: R2 = 0.238, P = 0.029), stress-70 protein (spot #107: R2 = 0.415, P = 0.0022), serine protease inhibitor A3K (spot #123: R2 = 0.392, P = 0.0031 and Spot #139: R2 = 0.4915, P = 0.0006), β-enolase (spot #379: R2 = 0.2668, P = 0.0197 and spot #341: R2 = 0.227, P = 0.034), enoyl-CoA hydratase (spot #762: R2 = 0.204, P = 0.046) and fumarate hydratase (spot #423: R2 = 0.211, P = 0.042 and spot #426: R2 = 0.266, P = 0.02).
4. Discussion
DIGE analysis revealed that there is little or no sexual dimorphism in the muscle response to artificial selection on running capacity. Significant main effects of sex (15 spots) and strain (40 spots) were clearly evident in the soleus proteome but just 1 spot (#379; β-enolase) exhibited a statistically significant sex strain interaction. As anticipated, the myofibre profile was identical in HCR and LCR soleus and HCR muscle exhibited significantly greater mitochondrial content (Table 2). Indeed, the majority of differences between the HCR and LCR soleus proteomes were of mitochondrial origin, which support recent hypothesis-led studies [8,37] and our preliminary proteome profiling [11] using label-free LC–MS. Furthermore, our current DIGE analysis highlighted mitochondrial protein import as a potential mechanism underpinning the enhanced mitochondrial function in HCR muscle.
Further to the conventional analysis of the DIGE data, we used conditional independence mapping as an objective means to identify associations between protein species that exhibited significant differences in abundance (Fig. 3). This novel approach highlighted important interactions, which were not obvious from first-principle deduction of gene ontology annotations. The 2most prominent networks highlighted by conditional independence mapping centred on protein disulphide isomerase A3 (PDIA3; spot #210) and 60 kDa heat shock protein (CH60; spot #238). This places particular emphasis on these proteins as candidate biomarkers and potential mediators of the differences between HCR and LCR skeletal muscle.
4.1. PDIA3 species are associated with STAT3 signalling
PDIA3, also known as endoplasmic reticulum protein 57 (ERp57) or glucose-responsive protein58 (GRP58), was first described as a thiol-dependent reductase [38,39]. However, silencing of PDIA3 does not have major effects on ER morphology or processes such as the unfolded protein response, but instead significantly affects STAT3 signalling [40]. Accordingly, PDIA3 and STAT3 are reported to interact in a ~200–400 kDa ‘statosome’, which is capable of influencing cytokine (IL-6) signalling in hepatoma (Hep3B) cells [41]. In fibroblasts, the protein abundance, Y705 phosphorylation and transcriptional activity of STAT3 are augmented in the absence of PDIA3 and this effect is reversed by transfection with PDIA3 directed to the ER lumen [40]. Changes in the abundance of PDIA3 have been reported due to ageing or exercise. PDIA3 is more abundant in the gastrocnemius of elderly compared to young rats [42], and endurance training is associated with an increase in PDIA3 abundance in human muscle mitochondria [43]. However, the current DIGE data depicts changes across different species of PDIA3 rather than differences in total abundance.
We resolved PDIA3 as 4 spots (220, 206, 210 and 202) of equivalent Mr but different pI indicating different protein species, which may be due to post-translational modifications or sequence variations. A similar PDIA3 spot pattern has been reported in hepatic tissue [44], where some PDIA3 species exhibited diurnal fluctuations. PDIA3 spot 4 and spot 3 reported in [44] were found to contain phosphorylated (S150) PDIA3 and probably correspond to spots 220 and 206 of current DIGE map, which were more abundant in LCR muscle. Our proteome mining work failed to unambiguously identify site-specific phosphorylations of PDIA3 but Kita et al. [44] used a phosphorylation specific Ab to show that S150 phosphorylation of PDIA3 occurs in response to periods of fasting greater than 12 h [44]. In our experiment rats were fasted overnight (~12 h) so the significant differences in muscle PDIA3 spot pattern (Table 4) may also be a response to fasting. Nevertheless, the period of fasting was standardised across male and female, HCR and LCR groups, meaning LCR exhibit a greater response than HCR to an equivalent metabolic stress. Kita et al. [44] report that native species of PDIA3 co-precipitate with STAT3 whereas S150 phosphorylation of PDIA3 blocks this interaction. If S150 phosphorylation of PDIA3 is also greater in LCR soleus then selection on low running capacity might be associated with greater STAT3 signalling. In support of this suggestion, the STAT3 pathway was the most highly-enriched gene-set in LCR gastrocnemius [10], and unsupervised bibliometric analysis highlighted robust connection between STAT3 and PDIA3 (Fig. 4). Using western blotting (Fig. 5), we found no differences in STAT3 protein abundance or Y705 phosphorylation but S727 phosphorylation of STAT3 was greater in LCR soleus.
The canonical STAT3 pathway is activated by cytokine receptors (e.g. leptin and IL-6) and involves tyrosine (Y705) phosphorylation of STAT3 by Janus kinases [45]. Tyrosine phosphorylation enables homo-dimerisation and nuclear translocation of STAT3, whereas serine (S727) phosphorylation of STAT3 disrupts this process [46]. S727 phosphorylation may be mediated by ERK-family mitogen activated protein kinases or ERK-independent processes involving the mammalian target of rapamycin (mTOR). Interestingly mTOR was one of the genes listed in the STAT3 pathway enriched in LCR muscle transcriptome [10]. Moreover, PDIA3 also interacts with mTOR and may be mechanistically involved in the activation of mTOR by oxidants such as phenylarsine oxide [47]. Phosphorylation of Akt S473 by mTOR is prevented when cells are incubated with a 2-fold excess of amino acids. This effect co-occurs with S727 phosphorylation of STAT3 while deletion of STAT3 abolishes the inhibitory effects of amino acids on insulin signalling. Using a STAT3 reporter assay, [48] reports that amino acid excess increases the transcriptional activity of STAT3 and leads to greater expression of SOCS3, which may inhibit insulin signalling. These effects in hepatocytes could also be seen in muscle and adipose cells, therefore, S727 phosphorylation of STAT3 in LCR soleus may contribute to the greater insulin resistance of LCR animals and warrant further investigation.
Amongst other proteins in the PDIA3 cluster were serine protease inhibitor A3L (Serpin; Spot #539), which has a STAT3 site in its promoter and exhibited the most prominent increase (3.18-fold greater) in LCR muscle, and N-myc down regulated gene 2 (NDRG2; spot #433), which is reported to modulate STAT3 and SOCS3 responses to cytokine stimulation in U937 lymphoblasts [49]. NDRG2 has previously been associated with modulation of insulin signalling [50] and can be phosphorylated at T330, S332 and T348 by serum- and glucocorticoid-induced kinase 1 (SGK1) [51], at S332 by PKCθ and at T348 by Akt [50]. Evidence of phosphorylation at each of these sites was detected during proteome mining but it was not possible to distinguish spots wherein phosphorylation patterns were specific to an individual kinase. Nevertheless, interrogation of CID and ETD spectra revelled spots #431 and #433, which were more abundant in LCR, lacked any evidence of phosphorylation at S338, which is a commonly reported modification to NDRG2 (131 MS reports in PhosphoSite) that was present in the other NDRG2 spots. Currently no empirical evidence of an upstream kinase exists but NetPhosK predicted casein kinase 1 (CK1) may target S338 of NDRG2. CK1 has been implicated with metabolic circadian rhythms via its modulation of the degradation rate of PGC-1α [52]. Therefore, greater phosphorylation of NDRG2 S338 may be consistent with the lesser mitochondrial content or greater response to fasting in LCR.
Fumarate hydratase was also a prominent component of the PDIA3 cluster (Fig. 2B), and was resolved as 3 spots that were significantly less abundant in LCR muscle (Table 4). The lesser overall abundance of fumarate hydratase provides protein-level confirmation of earlier transcriptome data [10] and may point to alterations in amino acid or purine metabolism. Fumarate hydratase is an anaplerotic entry point for phenylalanine and tyrosine to the TCA cycle and is also associated with purine metabolism [53]. Accordingly we found that several other enzymes of purine metabolism were less abundant in LCR, including 3-mercaptopyruvate sulfurtransferase and hypoxanthine– guanine phosphoribosyltransferase. Alongside its role in purine metabolism, 3-mercaptopyruvate sulfurtransferase performs the final reaction in the metabolism of glycine, serine and threonine to produce pyruvate. A lesser capacity for purine metabolism could negatively effect muscle energy regulation. In addition, severe fumarase deficiency may inhibit fatty acid oxidation due to inadequate production of oxaloacetone, which is required to accept acetyl-CoA in to the TCA cycle. This latter effect seems consistent with the lesser abundance of ACADV and enoyl-CoA hydratase, which were also clustered with PDIA3 spot #210 (Fig. 2B).
4.2. CH60 is a key node for mitochondrial proteins
The second most prominent cluster is centred on 60 kDa heat shock protein (spot #238). This finding likely relates to the differences in mitochondrial content between HCR and LCR muscle, which are widely acknowledged. The mitochondrial proteome encompasses approximately 1000 known proteins [54], many of which have been described in proteome mining studies of muscle mitochondria (e.g. [55]). The majority of mitochondrial proteins are transcribed from nuclear genes whereas 13 are derived from mitochondrial DNA [56]. Nuclear-encoded proteins often contain target sequences to direct the pre-protein to mitochondria. This process is mediated by cytosolic chaperones including heat shock cognate 71 kDa protein [57], which was more abundant in HCR soleus. Chaperone binding prevents aggregation of the precursor and facilitates its recognition by the translocase of the outer membrane (TOM), which is the first step in mitochondrial protein import. Proteins destined for the mitochondrial matrix must also transverse the inner membrane, and this energy-dependent process is driven by stress-70 protein (more commonly known as mitochondrial heat shock protein 70; mtHsp70). Within the matrix pre-sequences are cleaved and nascent proteins are folded in to their mature form by the chaperonin complex, which is a heteroligomeric structure consisting of subunits of 60 kDa heat shock protein (chaperonin 60; CH60) and 10 kDa heat shock protein (chaperonin 10; CH10). The crucial role of mitochondrial protein import is illustrated by data from yeast [58] reporting that 5 genes are essential for mitochondrial viability, and the products of each of these genes are components of the mitochondrial protein import machinery. Included in this list are chaperonin 60 and stress protein-70, which were significantly less abundant in LCR soleus (Table 4). In addition to being lethal in yeast, mutation of CH60 is related with inherited human diseases, including spastic paraplegia and hypomyelinating leukodystrophy [59].
It is important to mention that we detected CH60 in 5 spots and just one of these spots was more abundant in HCR muscle. This is consistent with our previous proteomics data [2] in human skeletal muscle where 3 species of CH60 were detected and one spot was significantly more abundant in samples collected after high-intensity interval training. As yet the individual CH60 species have not been characterised but it is clear that innate and acquired differences in muscle aerobic capacity affect proteins such as CH60 at the species level, in addition to reported changes in the total abundance of the protein (e.g. [60]). It remains to be determined whether the different species of CH60 influence the oligomeric structure or activity of the chaperonin complex. Similar to CH60, stress-70 protein was detected as a series of 3 spots, one of which (spot #53) was more abundant in HCR muscle. Consistent with our findings (Table 4) using artificial selection and earlier experience in exercise training [61], chronic low-frequency stimulation (CLFS) also increases Stress-70 protein and CH60 [60]. In response to CLFS, the magnitude of the change in abundance of Stress-70 and CH60 is proportionally greater than that of respiratory chain proteins such as COX [60], which suggests that robust augmentation of protein import is necessary to facilitate mitochondrial adaptations.
Consistent with our previous findings [11], several of the mitochondrial proteins that exhibited differences between HCR and LCR (Table 4) were also highlighted in the ‘longevity interactome’ produced from genes that modify lifespan when mutated in model organisms such as yeast and Caenorhabditis elegans [62]. In particular, proteins involved in mitochondrial protein import (heat shock cognate 71, stress-70 protein and 60 kDa heat shock protein) fatty acid metabolism (aspartate aminotransferase) and NDRG2 belong to the ‘core set’ of the longevity interactome, which comprises genes that also exhibit significant differences between muscle of young and elderly humans [62]. Therefore, our current data (Table 4) in young animals may link with the differences in natural ageing and life expectancy of HCR and LCR rats [63].
In LCR animals, serumleptin concentrations were negatively correlated with the majority of metabolic proteins that were differentially expressed between HCR and LCR muscle. Typically, leptin signalling stimulates muscle fatty acid oxidation and glucose uptake and improves insulin sensitivity [61]. We found that serum leptin levels were greater in males than females and male LCR exhibited the highest serum leptin concentrations. These findings extend earlier work in females [36] and the inverse correlations between serum leptin and the muscle proteome data suggest that LCR animals are leptin resistant.
5. Conclusion
The current work reports the first broad-scale profiling of the muscle proteome from male and female animals selected based on either high- or low-running capacity. Relatively few differences were detected between the soleus proteome of male and female animals, and sex-specific protein regulation (Table 3) did not demonstrate strong functional clustering. In contrast, selection on running capacity was associated with differences in functional groups, such as glycolysis, TCA cycle and oxidative phosphorylation. Importantly, graphical network analysis of the proteome data highlighted potential interactions which were not obvious from first-principle deduction of the gene ontology information. The application of conditional independence mapping enabled us to gain insight to latent interactions, which may highlight underlying mechanisms. The utility of this novel unsupervised approach to the interpretation of DIGE data warrants wider application, and our current work also supports the use of bibliometric analysis as an unbiased tool for discovering potential protein networks. In particular, we found PDIA3 and CH60 to be key nodes associated with differences in muscle aerobic capacity and our data further highlight STAT3 and NDRG2 as novel candidate mechanisms, which may underly the differences in muscle insulin sensitivity.
A reference 2D gel image is available at the World 2D-PAGE repository (http://world-2dpage.expasy.org/repository/), accession number 0069. Spot numbers used throughout this manuscript are consistent with the World-2DPAGE reference map and each spot coordinate is linked to the mass spectrometry protein identification. The associated CID and ETD MS/MS ions data in Mascot generic format are available upon request. Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.jprot.2014.04.015.
Supplementary Material
Biological significance.
Quantitative proteome profiling revealed that there is little or no sexual dimorphism in the skeletal muscle response to artificial selection on running capacity. Instead we found that noncanonical STAT3 signalling may be associated with low exercise capacity and skeletal muscle insulin resistance. Importantly, this discovery was made using unsupervised multivariate association mapping and bibliometric network analyses. This allowed our interpretation of the findings to be guided by patterns within the data rather than our preconceptions about which proteins or processes are of greatest interest. Moreover, we demonstrate that this novel approach can be applied to 2D gel analysis, which is unsurpassed in its ability to profile protein species but currently has few dedicated bioinformatic tools.
Acknowledgements
This work was supported by the Royal Society (JGB, LGK and SLB) (Grant no. RG080544). Y-W Chen was partially supported by DOD W81XWH-10-1-0659, NIH/NIAMS 1RO1AR052027 and NIH/NICHD 1R24HD05084.
The LCR-HCR rat model system was funded by the National Center for Research Resources grant R24RR017718 and is currently supported by the Office of Research Infrastructure Programs/OD grant R24OD010950 (LGK and SLB) from the National Institutes of Health of the United States. SLB was also supported by National Institutes of Health grants RO1 DK077200 and R01GM104194. We acknowledge the expert care of the rat colony provided by Molly Kalahar and Lori Gilligan. Contact LGK (lgkoch@med.umich.edu) or SLB (brittons@umich.edu) for information on the LCR and HCR rats: these rat models are maintained as an international collaborative resource at the University of Michigan, Ann Arbor.
Footnotes
Conflict of interest
We have no conflicts of interest to disclose.
REFERENCES
- 1.Warburton DE, Nicol CW, Bredin SS. Health benefits of physical activity: the evidence. Can Med Assoc J. 2006;174:801–809. doi: 10.1503/cmaj.051351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holloway KV, O'Gorman M, Woods P, Morton JP, Evans L, Cable NT, et al. Proteomic investigation of changes in human vastus lateralis muscle in response to interval-exercise training. Proteomics. 2009;9:5155–5174. doi: 10.1002/pmic.200900068. [DOI] [PubMed] [Google Scholar]
- 3.Burniston JG. Adaptation of the rat cardiac proteome in response to intensity-controlled endurance exercise. Proteomics. 2009;9:106–115. doi: 10.1002/pmic.200800268. [DOI] [PubMed] [Google Scholar]
- 4.Burniston JG. Changes in the rat skeletal muscle proteome induced by moderate-intensity endurance exercise. Biochim Biophys Acta Protein Proteomics. 2008;1784:1077–1086. doi: 10.1016/j.bbapap.2008.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Koch LG, Britton SL. Artificial selection for intrinsic aerobic endurance running capacity in rats. Physiol Genomics. 2001;5:45–52. doi: 10.1152/physiolgenomics.2001.5.1.45. [DOI] [PubMed] [Google Scholar]
- 6.Wisloff U, Najjar SM, Ellingsen O, Haram PM, Swoap S, Al-Share Q, et al. Cardiovascular risk factors emerge after artificial selection for low aerobic capacity. Science. 2005;307:418–420. doi: 10.1126/science.1108177. [DOI] [PubMed] [Google Scholar]
- 7.Rivas DA, Lessard SJ, Saito M, Friedhuber AM, Koch LG, Britton SL, et al. Low intrinsic running capacity is associated with reduced skeletal muscle substrate oxidation and lower mitochondrial content in white skeletal muscle. Am J Physiol Regul Integr Comp Physiol. 2011;300:R835–R843. doi: 10.1152/ajpregu.00659.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seifert EL, Bastianelli M, Aguer C, Moffat C, Estey C, Koch LG, et al. Intrinsic aerobic capacity correlates with inherent mitochondrial oxidative and H2O2 emission capacities without major shifts in myosin heavy chain isoform. J Appl Physiol. 2012;113:1624–1634. doi: 10.1152/japplphysiol.01475.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lessard SJ, Rivas DA, Stephenson EJ, Yaspelkis BB, Koch LG, et al. Exercise training reverses impaired skeletal muscle metabolism induced by artificial selection for low aerobic capacity. Am J Physiol Regul Integr Comp Physiol. 2010;300:R175–R182. doi: 10.1152/ajpregu.00338.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kivelä R, Silvennoinen M, Lehti M, Rinnankoski-Tuikka R, Purhonen T, et al. Gene expression centroids that link with low intrinsic aerobic exercise capacity and complex disease risk. FASEB J. 2010;24:4565–4574. doi: 10.1096/fj.10-157313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malik Z, Cobley J, Morton J, Close G, Edwards B, Koch LG, et al. Label-free LC–MS profiling of skeletal muscle reveals heart-type fatty acid binding protein as a candidate biomarker of aerobic capacity. Proteomes. 2013;1:290–308. doi: 10.3390/proteomes1030290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jungblut P, Holzhütter H, Apweiler R, Schlüter H. The speciation of the proteome. Chem Cent J. 2008;2:1–10. doi: 10.1186/1752-153X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Højlund K, Yi Z, Hwang H, Bowen B, Lefort N, Flynn CR, et al. Characterization of the human skeletal muscle proteome by one-dimensional gel electrophoresis and HPLC-ESI- MS/MS. Mol Cell Proteomics. 2008;7:257–267. doi: 10.1074/mcp.M700304-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker KC, Walsh R, Salajegheh M, Amato A, Krastins B, Sarracino D, et al. Characterization of human skeletal muscle biopsy samples using shotgun proteomics. J Proteome Res. 2009;8:3265–3277. doi: 10.1021/pr800873q. [DOI] [PubMed] [Google Scholar]
- 15.Dang X, Scotcher J, Wu S, Chu RK, Tolić N, et al. The first pilot project of the consortium for top down proteomics: a status report. Proteomics. 2014 doi: 10.1002/pmic.201300438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burniston JG, Hoffman EP. Proteomic responses of skeletal and cardiac muscle to exercise. Expert Rev Proteomics. 2011;8:361–377. doi: 10.1586/epr.11.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burniston JG, Kenyani J, Wastling JM, Burant CF, Qi NR, Koch LG, et al. Proteomic analysis reveals perturbed energy metabolism and elevated oxidative stress in hearts of rats with inborn low aerobic capacity. Proteomics. 2011;11:3369–3379. doi: 10.1002/pmic.201000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kislinger T, Gramolini AO, Pan Y, Rahman K, MacLennan DH, Emili A. Proteome dynamics during C2C12 myoblast differentiation. Mol Cell Proteomics. 2005;4:887–901. doi: 10.1074/mcp.M400182-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Bacciu D, Etchells TA, Lisboa PJG, Whittaker J. Efficient identification of independence networks using mutual information. Comput Stat. 2013;28:621–646. [Google Scholar]
- 20.Maher ACAA, Vockley MA, Tarnopolsky JA, Mark M. Women have higher protein content of β-oxidation enzymes in skeletal muscle than men. PLoS One. 2010;5:e12025. doi: 10.1371/journal.pone.0012025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hart N, Sarga L, Csende Z, Koltai E, Koch LG, Britton Sl, et al. Resveratrol enhances exercise training responses in rats selectively bred for high running performance. Food Chem Toxicol. 2013;61:53–59. doi: 10.1016/j.fct.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tweedie C, Romestaing C, Burelle Y, Safdar A, Tarnopolsky MA, Seadon S, et al. Lower oxidative DNA damage despite greater ROS production in muscles from rats selectively bred for high running capacity. Am J Physiol Regul Integr Comp Physiol. 2010;300:R545–R553. doi: 10.1152/ajpregu.00250.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noland RC, Thyfault JP, Henes ST, Whitfield BR, Woodlief TL, Evans JR, et al. Artificial selection for high-capacity endurance running is protective against high-fat diet-induced insulin resistance. Am J Physiol Endocrinol Metab. 2007;293:E31–E41. doi: 10.1152/ajpendo.00500.2006. [DOI] [PubMed] [Google Scholar]
- 24.Oh TS, Choi JW, Choi DK, Mukherjee R, Liu H, Yun JW. Gender dimorphism in skeletal muscle proteome between lean and diet-induced obese rats. Cell Physiol Biochem. 2011;28:981–996. doi: 10.1159/000335811. [DOI] [PubMed] [Google Scholar]
- 25.Choi M, Choi JW, Chaudhari HN, Aseer KR, Mukherjee R, Yun JW. Gender-dimorphic regulation of skeletal muscle proteins in streptozotocin-induced diabetic rats. Cell Physiol Biochem. 2013;31:408–420. doi: 10.1159/000343378. [DOI] [PubMed] [Google Scholar]
- 26.Metskas LA, Kulp M, Scordilis SP. Gender dimorphism in the exercise-naïve murine skeletal muscle proteome. Cell Mol Biol Lett. 2010;15:507–516. doi: 10.2478/s11658-010-0020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamadeh MJ, Devries MC, Tarnopolsky MA. Estrogen supplementation reduces whole body leucine and carbohydrate oxidation and increases lipid oxidation in men during endurance exercise. J Clin Endocrinol Metab. 2005;90:3592–3599. doi: 10.1210/jc.2004-1743. [DOI] [PubMed] [Google Scholar]
- 28.Burniston JG, McLean L, Beynon RJ, Goldspink DF. Anabolic effects of a non-myotoxic dose of the β2-adrenergic receptor agonist clenbuterol on the rat plantaris muscle. Muscle Nerve. 2007;35:217–223. doi: 10.1002/mus.20684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 31.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoffmann R, Valencia A. A gene network for navigating the literature. Nat Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- 33.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 34.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong HY, Yoo GS, Choi JK. Direct Blue 71 staining of proteins bound to blotting membranes. Electrophoresis. 2000;21:841–845. doi: 10.1002/(SICI)1522-2683(20000301)21:5<841::AID-ELPS841>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 36.Novak CM, Escande C, Burghardt PR, Zhang M, Barbosa MT, Chini EN, et al. Spontaneous activity, economy of activity, and resistance to diet-induced obesity in rats bred for high intrinsic aerobic capacity. Horm Behav. 2010;58:355–361. doi: 10.1016/j.yhbeh.2010.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stephenson EJ, Stepto NK, Koch LG, Britton SL, Hawley JA. Divergent skeletal muscle respiratory capacities in rats artificially selected for high and low running ability: a role for Nor1? J Appl Physiol. 2012;113:1403–1412. doi: 10.1152/japplphysiol.00788.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirano N, Shibasaki F, Sakai R, Tanaka T, Nishida J, Yazaki Y, et al. Molecular cloning of the human glucose-regulated protein ERp57/GRP58, a thiol-dependent reductase. Identification of its secretory form and inducible expression by the oncogenic transformation. Eur J Biochem. 1995;234:336–342. doi: 10.1111/j.1432-1033.1995.336_c.x. [DOI] [PubMed] [Google Scholar]
- 39.Frickel EM, Frei P, Bouvier M, Stafford WF, Helenius A, et al. ERp57 is a multifunctional thiol-disulfide oxidoreductase. J Biol Chem. 2004;279:18277–18287. doi: 10.1074/jbc.M314089200. [DOI] [PubMed] [Google Scholar]
- 40.Coe H, Jung J, Groenendyk J, Prins D, Michalak M. ERp57 modulates STAT3 signaling from the lumen of the endoplasmic reticulum. J Biol Chem. 2010;285:6725–6738. doi: 10.1074/jbc.M109.054015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo GG, Patel K, Kumar V, Shah M, Fried VA, Etlinger D, et al. Association of the chaperone glucose-regulated protein 58 (GRP58/ER-60/ERp57) with Stat3 in cytosol and plasma membrane complexes. J Interferon Cytokine Res. 2002;22:555–563. doi: 10.1089/10799900252982034. [DOI] [PubMed] [Google Scholar]
- 42.Piec I, Listrat A, Alliot J, Chambon C, Taylor RG, Bechet D. Differential proteome analysis of aging in rat skeletal muscle. FASEB J. 2005;19:1143–1145. doi: 10.1096/fj.04-3084fje. [DOI] [PubMed] [Google Scholar]
- 43.Egan B, Dowling P, O'Connor PL, Henry M, Meleady P, Zierath JR, et al. 2-D DIGE analysis of the mitochondrial proteome from human skeletal muscle reveals time course-dependent remodelling in response to 14 consecutive days of endurance exercise training. Proteomics. 2011;11:1413–1428. doi: 10.1002/pmic.201000597. [DOI] [PubMed] [Google Scholar]
- 44.Kita K, Okumura N, Takao T, Watanabe M, Matsubara T, Nishimura O, et al. Evidence for phosphorylation of rat liver glucose-regulated protein 58, GRP58/ERp57/ER-60, induced by fasting and leptin. FEBS Lett. 2006;580:199–205. doi: 10.1016/j.febslet.2005.11.074. [DOI] [PubMed] [Google Scholar]
- 45.Kaptein A, Paillard V, Saunders M. Dominant negative stat3 mutant inhibits interleukin-6-induced Jak-STAT signal transduction. J Biol Chem. 1996;271:5961–5964. doi: 10.1074/jbc.271.11.5961. [DOI] [PubMed] [Google Scholar]
- 46.Chung J, Uchida E, Grammer TC, Blenis J. STAT3 serine phosphorylation by ERK-dependent and -independent pathways negatively modulates its tyrosine phosphorylation. Mol Cell Biol. 1997;17:6508–6516. doi: 10.1128/mcb.17.11.6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramírez-Rangel I, Bracho-Valdés I, Vázquez-Macías A, Carretero-Ortega J, Reyes-Cruz G, Vázquez-Prado J. Regulation of mTORC1 complex assembly and signaling by GRp58/ERp57. Mol Cell Biol. 2011;31:1657–1671. doi: 10.1128/MCB.00824-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim JH, Yoon MS, Chen J. Signal transducer and activator of transcription 3 (STAT3) mediates amino acid inhibition of insulin signaling through serine 727 phosphorylation. J Biol Chem. 2009;284:35425–35432. doi: 10.1074/jbc.M109.051516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee EB, Kim A, Kang K, Kim H, Lim JS. NDRG2-mediated modulation of SOCS3 and STAT3 activity inhibits IL-10 production. Immune Netw. 2010;10:219–229. doi: 10.4110/in.2010.10.6.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Burchfield JG, Lennard AJ, Narasimhan S, Hughes WE, Wasinger VC, Corthalis GL, et al. Akt mediates insulin-stimulated phosphorylation of Ndrg2: evidence for cross-talk with protein kinase C theta. J Biol Chem. 2004;279:18623–18632. doi: 10.1074/jbc.M401504200. [DOI] [PubMed] [Google Scholar]
- 51.Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, et al. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li S, Chen XW, Yu L, Saltiel AR, Lin JD. Circadian metabolic regulation through crosstalk between casein kinase 1δ and transcriptional coactivator PGC-1α. Mol Endocrinol. 2011;25:2084–2093. doi: 10.1210/me.2011-1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Des Rosiers C, Labarthe F, Lloyd SG, Chatham JC. Cardiac anaplerosis in health and disease: food for thought. Cardiovasc Res. 2011;90:210–219. doi: 10.1093/cvr/cvr055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Smith AC, Blackshaw JA, Robinson AJ. MitoMiner: a data warehouse for mitochondrial proteomics data. Nucleic Acids Res. 2012;40:D1160–D1167. doi: 10.1093/nar/gkr1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lefort N, Yi Z, Bowen B, Glancy B, De Filippis EA, Mapes R, et al. Proteome profile of functional mitochondria from human skeletal muscle using one-dimensional gel electrophoresis and HPLC-ESI-MS/MS. J Proteomics. 2009;72:1046–1060. doi: 10.1016/j.jprot.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anderson S, Bankier T, Barrell G, de Bruijn MHL, Coulson R, Drouin J, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 57.Zara V, Ferramosca A, Robitaille-Foucher P, Palmieri F, Young JC. Mitochondrial carrier protein biogenesis: role of the chaperones Hsc70 and Hsp90. Biochem J. 2009;419:369–375. doi: 10.1042/BJ20082270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Baker KP, Schatz G. Mitochondrial proteins essential for viability mediate protein import into yeast mitochondria. Nature. 1991;349:205–208. doi: 10.1038/349205a0. [DOI] [PubMed] [Google Scholar]
- 59.Christensen JH, Nielsen MN, Hansen J, Füchtbauer A, Füchtbauer EM, West M, et al. Inactivation of the hereditary spastic paraplegia-associated Hspd1 gene encoding the Hsp60 chaperone results in early embryonic lethality in mice. Cell Stress Chaperones. 2010;15:851–863. doi: 10.1007/s12192-010-0194-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ornatsky OI, Connor MK, Hood DA. Expression of stress proteins and mitochondrial chaperonins in chronically stimulated skeletal muscle. Biochem J. 1995;311(Pt 1):119–123. doi: 10.1042/bj3110119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dube JJ, Bhatt BA, Dedousis N, Bonen A, O'Doherty RM. Leptin, skeletal muscle lipids, and lipid-induced insulin resistance. Am. J Physiol Regul Integr Comp Physiol. 2007;293:R642–R650. doi: 10.1152/ajpregu.00133.2007. [DOI] [PubMed] [Google Scholar]
- 62.Bell R, Hubbard A, Chettier R, Chen D, Miller JP, Kapahi P, et al. A human protein interaction network shows conservation of aging processes between human and invertebrate species. PLoS Genet. 2009;5:e1000414. doi: 10.1371/journal.pgen.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koch LG, Kemi OJ, Qi N, Leng SX, Bijma P, Gilligan LJ, et al. Intrinsic aerobic capacity sets a divide for aging and longevity. Circ Res. 2011;109:1162–1172. doi: 10.1161/CIRCRESAHA.111.253807. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.