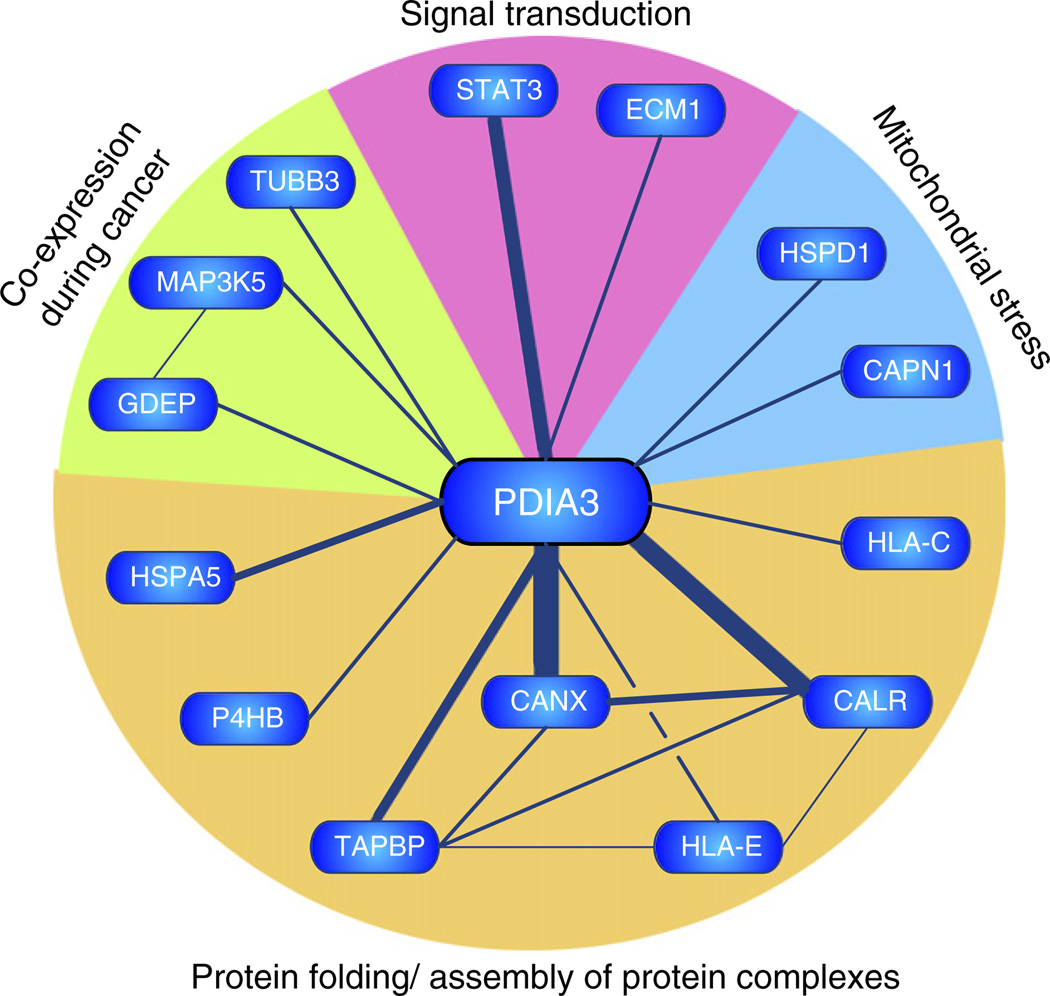
Fig. 4.
Bibliometric network of PDIA3. A gene model representing the bibliometric network of PDIA3 was constructed by automated text-mining (data redrawn from iHOP; information hyperlinked over proteins; www.ihop-net. org). Nodes (genes) are connected by edges, which represent co-occurrence within sentences of peer-reviewed published literature. Edge thickness approximates the number of supporting sentences. The majority of associations (CANX, calnexin; CALR, calreticulin, TAPBP, tapsin; HLA-E/HLA-C, major histocompatibility complex IE/C; HSPA5, glucose regulated protein 78; P4HB, prolyl-4-hydroxylase beta) were related to protein folding or assembly of protein complexes within the endoplasmic reticulum. PDIA3 was also reported to interact (TUBB3, class III beta tubulin) or be co-expressed with mitogen activated protein kinase kinase kinase 5 (MAP3K5) and gene differentially expressed in prostate (GDEP) in malignant stages of prostate cancer. Fourteen sentences reported interaction or colocalisation of PDIA3 with STAT3.