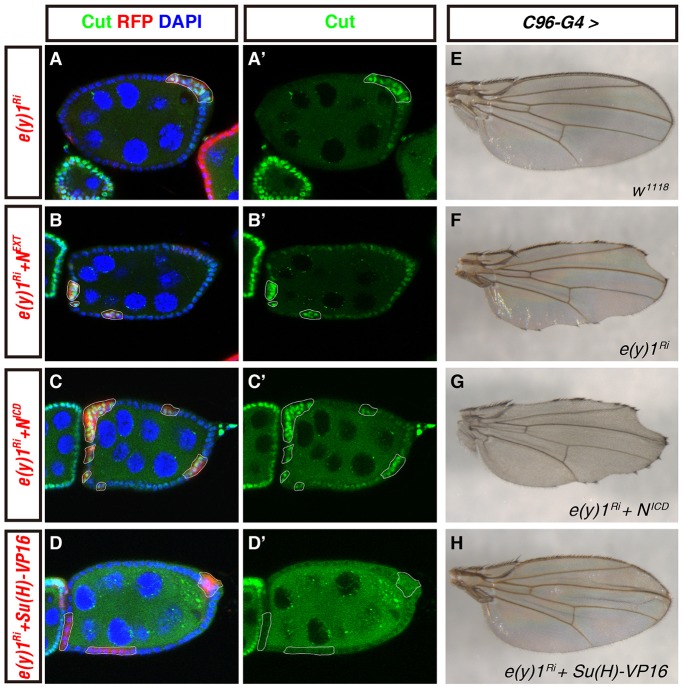
Fig. 4.
Epistatic analyses of E(y)1 in the Notch pathway in endocycling follicle cells and adult wings. (A–D′) E(y)1 regulates Notch signaling downstream of Notch proteolytic cleavage in mid-stage follicle cells. Cell nuclei were stained with DAPI (blue; A,B,C,D). Flip-out clones were marked by the expression of RFP (red; A,B,C,D) and outlined by white lines. Mid-stage egg chambers were stained for Cut in green (A–D′). Because a stau:GFP transgene was recombined in the hsFLP-carrying X chromosome, punctate green signals were detected within the germ cells. (A,B,C,D) Merged pictures of three channels; (A′,B′,C′,D′) Single channel of Cut staining. (A,A′) Follicle cell clones of e(y)1-RNAi in a mid-stage egg chamber showed Cut upregulation. (B–C′) Expression of NEXT (B,B′) or NICD (C,C′) failed to stop Cut upregulation caused by e(y)1 depletion. (D,D′) Induction of the strong Su(H)–VP16 activator prevented Cut expression in mid-stage e(y)1-RNAi follicle cells. (E–H) Ectopic expression of Su(H)–VP16 suppressed the notched wing margin defect caused by C96-Gal4-driven e(y)1 RNAi. (E) C96-Gal4 control adult flies had normal wing margins. (F) A typical loss of wing margin in C96-Gal4>e(y)1-RNAi flies. (G) Co-expression of NICD under the same C96-Gal4 driver did not suppress wing margin defects caused by e(y)1 RNAi. (H) Co-expression of Su(H)–VP16 substantially suppressed the wing margin loss resulting from C96-Gal4>e(y)1-RNAi. All wings were from male flies. Superscript Ri after a gene denotes RNAi against that gene.