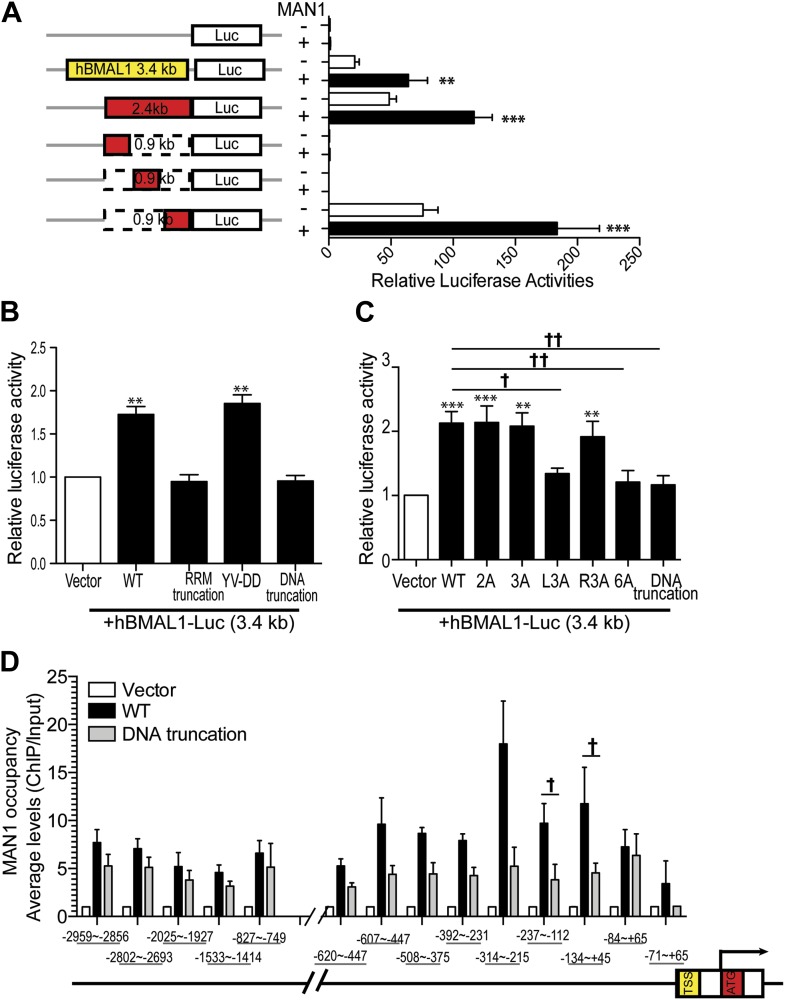
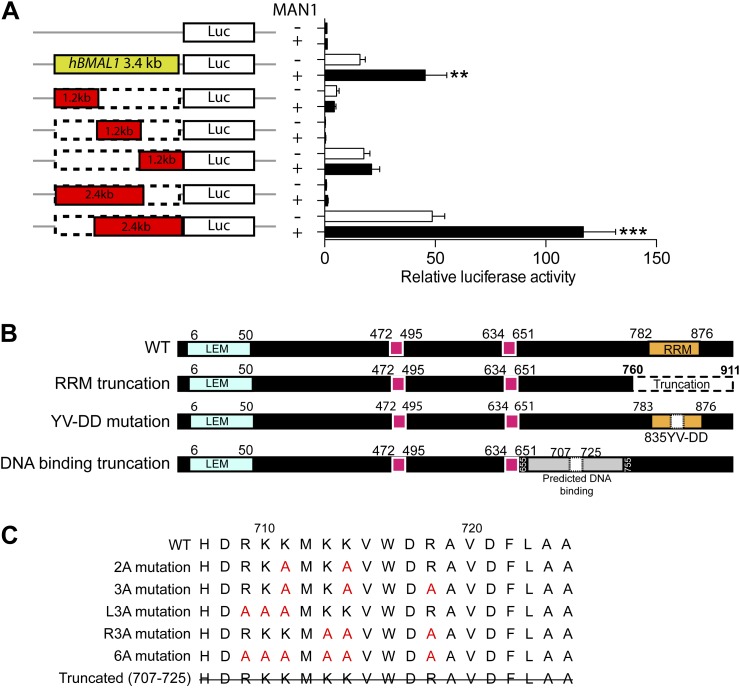
Figure 8. MAN1 binds to the BMAL1 promoter to enhance its transcription.
(A) Luciferase activities of deleted hBMAL1-promoter constructs in the absence or presence of MAN1 expression vectors. n = 3, Student's t test, **p < 0.01, ***p < 0.001. (B and C) Luciferase activities of the 3.4 Kb hBMAL1-Luc in the presence of MAN1 constructs as indicated. n = 3, **p < 0.01, ***p < 0.001 compared to control; †p < 0.05; ††p < 0.001 compared to WT MAN1. (D) ChIP analysis of MAN1 (WT or DNA binding truncation) for 14 segments of hBMAL1 promoter region. Data represent pull-down relative to input. n = 6, †p < 0.05, compared to WT MAN1. One-way ANOVA with Newman–Keuls test. All data are presented as ratio of means ± SEM.