Fig. 4.
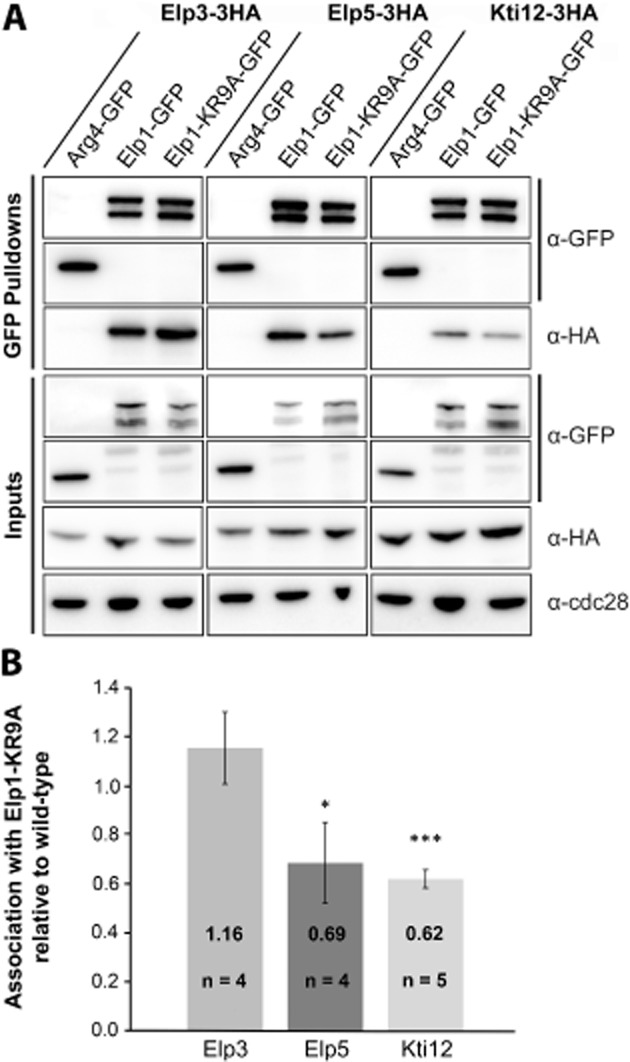
The elp1–KR9A mutation does not disrupt Elongator complex assembly but causes subtle differences in association with Elp5 and Kti12.
A. Western blot analysis of inputs and GFP–Trap immunoprecipitates of Elp1–GFP and Elp1–KR9A–GFP from strains expressing HA-tagged Elp3, Elp5 or Kti12. All samples were analysed with anti–GFP and anti-HA. Input samples were also analysed using anti-Cdc28 as a loading control. Arg4–GFP was confirmed to have similar pulldown efficiency to Elp1–GFP and was used as a control to demonstrate that interaction between Elp1 and the various HA tagged complex members was specific. All pulldown samples were analysed from the same blot. It should be noted that Elp1 is always observed by Western blot analysis as both a full-length and a faster migrating form (truncated at its N-terminus) that may be a degradation product or serve an as yet unknown function (Fichtner et al., 2003).
B. Quantification of co-immunoprecipitation efficiency. Immunoprecipitation of HA-tagged proteins was quantified by densitometry of the HA tag signals and normalized using the Elp1–GFP signals across the indicated number of replicates (n), setting the value for the wild-type strain in each case to 1.0. Error bars represent the standard deviation of the mean and the significance of the differences was analysed using a one way ANOVA, with ‘*’ indicating a P-value of < 0.05 (significant) and ‘***’ indicating a P-value of < 0.001 (very highly significant). Any small differences in Elp3 association were not statistically significant when analysed in this manner.
