Table 2. Sedimentation velocity data.
| sT,B | s20,wa | Mr (Da) | f/f0b | Axial ratio | |
|---|---|---|---|---|---|
| 22wtc | 1.47 ± 0.21 | 1.93 ± 0.27 | 8768 ± 1815 | 1.36 ± 0.17 | 2.1 |
| G1c | 1.49 ± 0.15 | 1.94 ± 0.20 | 7969 ± 1179 | 1.35 ± 0.12 | 2.0 |
| G2c | 1.72 ± 0.31 | 2.25 ± 0.41 | 10,938 ± 2924 | 1.17 ± 0.18 | ∼1 |
| G3c | 1.53 ± 0.19 | 2.00 ± 0.25 | 7850 ± 1445 | 1.32 ± 0.15 | 1.5 |
| G4c | 1.49 ± 0.15 | 1.95 ± 0.20 | 7853 ± 1198 | 1.35 ± 0.13 | 1.9 |
| G5c | 1.56 ± 0.28 | 2.03 ± 0.37 | 9695 ± 2568 | 1.29 ± 0.20 | 1.3 |
| G6c | 1.48 ± 0.19 | 1.93 ± 0.25 | 8036 ± 1489 | 1.36 ± 0.15 | 2.1 |
| G11c | 1.63 ± 0.28 | 2.13 ± 0.37 | 10,051 ± 2557 | 1.24 ± 0.18 | 1.3 |
| 26-merc | 1.08 ± 0.02 | 1.41 ± 0.15 | 8247 ± 1652 | 1.87 ± 0.05 | 8.6 |
| G1d | 0.62 ± 0.11 | 1.29 ± 0.23 | 7943 ± 1219 | 1.95 ± 0.29 | 14.3 |
| G5d | 0.65 ± 0.14 | 1.33 ± 0.32 | 7720 ± 1311 | 1.86 ± 0.32 | 12.6 |
| 22wtxe | 0.79 ± 0.13 | 1.24 ± 0.20 | 6813 ± 1556 | 1.98 ± 0.28 | 10.1 |
| 22wtxc | 1.50 ± 0.2 | 1.95 ± 0.26 | 6925 ± 1245 | 1.37 ± 0.18 | 2.1 |
| G5xe | 0.77 ± 0.09 | 1.20 ± 0.14 | 6541 ± 1064 | 2.05 ± 0.21 | 11.1 |
| G5xc | 1.54 ± 0.19 | 2.02 ± 0.25 | 7090 ± 1093 | 1.31 ± 0.12 | 1.4 |
as20,w values converted from sT,b values. Ranges are 95% confidence intervals (31).
bf/f0, buffer densities and viscosities were calculated using Sednterp (31), with 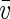 = 0.54 ml g−1 for G-quadruplex DNAs (32) and
= 0.54 ml g−1 for G-quadruplex DNAs (32) and 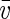 = 0.55 ml g−1 for the 26-mer ssDNA (36).
= 0.55 ml g−1 for the 26-mer ssDNA (36).
cIn 10 mM Tris–HCl (pH 8.0), 1 mM EDTA and 75 mM KCl.
dIn 10 mM Tris–HCl (pH 8.0), 1 mM EDTA and 75 mM TEA.
eIn 10 mM Tris–HCl (pH 8.0) and 1 mM EDTA (acid form).
