Figure 3.
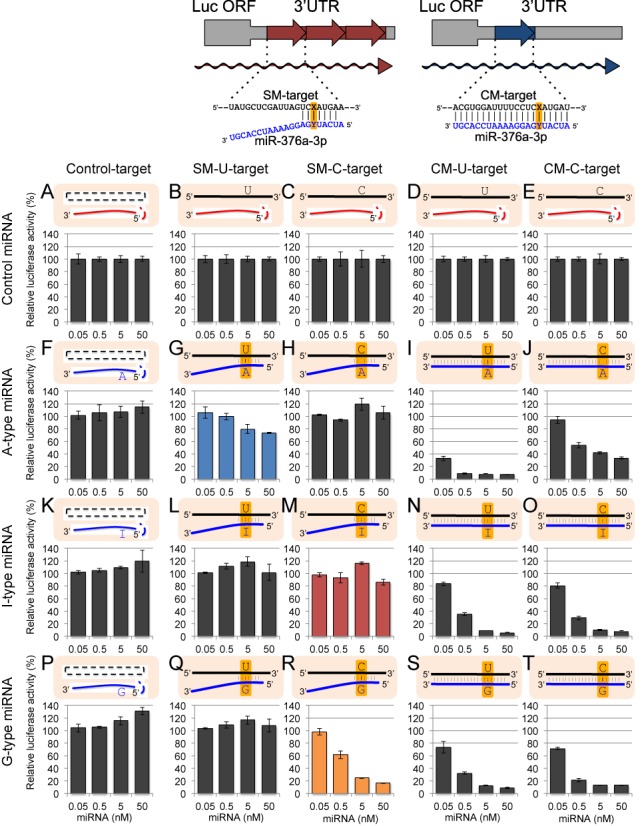
The results of reporter assays using miR-376a-2 duplex and its derivatives. The upper panels indicate the schematic structures of psiCHECK-SM-target and psiCHECK-CM-target constructs containing three tandem repeats of SM-target sequences and a CM-target sequence, respectively, in the 3′ UTR of Renilla luciferase gene of psiCHECK vector. Position Y indicates the possible editing site in the mRNA, and position X in the SM- or CM-target sequence indicates the position opposite to the the possible editing site of miRNA. Control siGY441 (A)–(E), wild-type A-type (F)–(J), I-type (K)–(O), or G-type (P)–(T) miR-376a-2 duplex was transfected with control psiCHECK-1 (A), (F), (K), (P), psiCHECK-SM-U-target (B), (G), (L), (Q), psiCHECK-SM-C-target (C), (H), (M), (R), psiCHECK-CM-U-target (D), (I), (N), (S), or psiCHECK-CM-C-target (E), (J), (O), (T), and pGL3-Control firefly luciferase expression construct were simultaneously transfected into HeLa cells. Relative luciferase activity was measured one day after transfection. Each miRNA was transfected at 0.05, 0.5, 5 and 50 nM, respectively. Complementarity between transfected miRNA and target construct is shown at top of each figure, and the possible editing sites were highlighted. Bar indicates the standard deviation of triplicated samples.
