Figure 1.
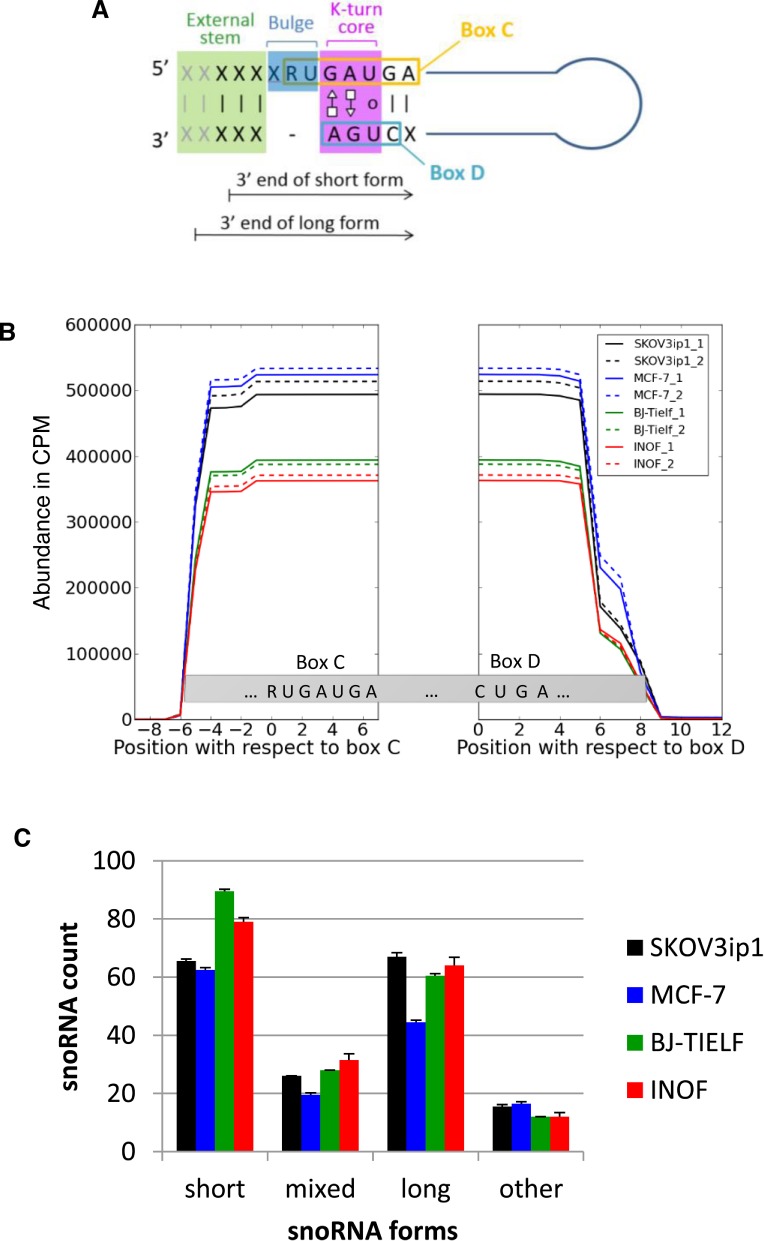
Comparison of box C/D snoRNA expression patterns in normal and cancer cell lines. (A) Schematic representation of box C/D snoRNA structure. Box C/D snoRNAs are small non-coding RNAs featuring two short sequence motifs (C: RUGAUGA and D: CUGA) that are aligned together through base pairing to form a characteristic structural k-turn motif. This motif typically involves a bulge upstream from the box C and non-canonical A–G and G–A base pairing between box C and D residues, preceded on the 5′ side by a stem involving canonical base pairing (12). X and R represent any nucleotide and purines, respectively. The 3′ end terminus of the short and long snoRNA forms detected in this study are indicated by arrows. (B) The processing pattern of box C/D snoRNA is conserved in normal and cancer cells. Sequence reads mapping to at least 77% of full-length box C/D snoRNAs in normal (BJ-Tielf, INOF), breast (MCF-7) and ovarian cancer cell lines (SKOV3ip) were counted and plotted with respect to their corresponding boxes C and D for every residue of all box C/D snoRNAs. CPM indicates count per million. All experiments were performed in duplicate. (C) Identification of two distinct forms of C/D snoRNA. Two general forms (long and short) of box C/D snoRNAs were identified according to the distance between their ends and their characteristic C/D motifs. The short forms (snoRNASH) start 4 or 5 nt upstream of their box C and end 2 or 3 nt downstream of their box D, while the long forms (snoRNAL) start 5 or 6 nt upstream of their box C and end 4 or 5 nt downstream of their box D. The number of snoRNAs displaying only short or only long forms, a mix of the two forms or neither long nor short forms (other) was counted in the different cell lines and presented in the form of a histogram. The standard deviation between the duplicate experiments is shown as error bars.
