Figure 3.
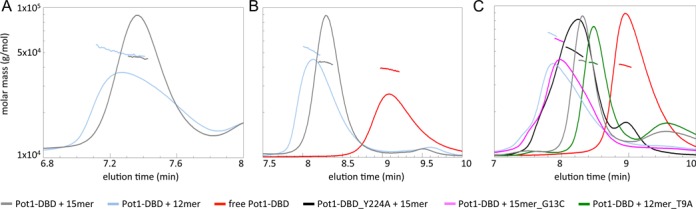
Pot1-DBD forms primarily a 1:1 complex at low concentrations and can form a larger complex at high concentrations in the presence of 12mer, but not 15mer. Amino acid mutation or base substitution can switch between these two binding modes. (A) RI traces (lines) and molar masses calculated by MALS (markers) of 34 μM Pot1-DBD+15mer and 34 μM Pot1-DBD+12mer complexes in 20 mM Tris–HCl pH 8.0, 50 mM NaCl. (B) Increasing the concentration of NaCl to 400 mM causes Pot1-DBD+12mer to elute earlier than Pot1-DBD+15mer and with a variable but larger average molar mass. Protein and DNA are both at 34 μM. (C) Increasing the concentration of Pot1-DBD to 170 μM and increasing the molar ratio to 2:1, Pot1-DBD:oligonucleotide pushes the 12mer-binding mode toward the 2:1 complex. This 12mer-binding mode can be forced to form a 1:1 complex upon 12mer_T9A substitution. Conversely, Pot1-DBD_Y224A mutation or 15mer_G13C substitution can switch the 15mer-binding mode to the 12mer-binding mode. All complexes were injected at 170 μM protein + 85 μM DNA.
