Figure 5.
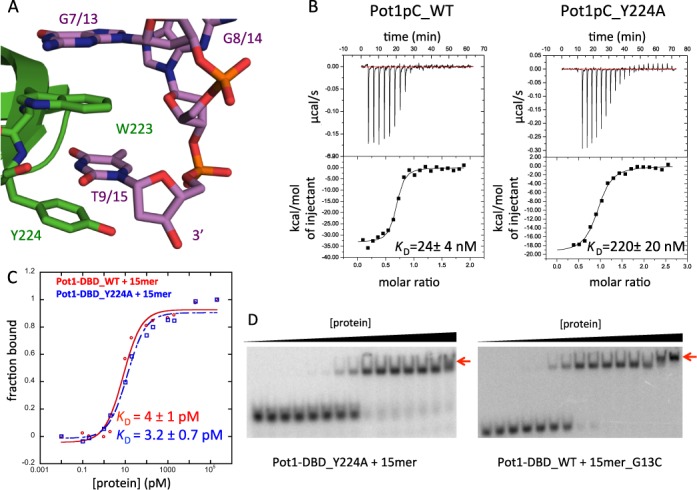
Disruption of the interaction between Pot1pC and the 3′ end of 15mer switches Pot1-DBD to the 12mer-binding mode. (A) In the Pot1pC+9mer complex, nucleotides G7 and T9 of 9mer (corresponding to G13 and T15 in 15mer) form an extended aromatic stack with Trp223 and Tyr224 of Pot1pC/Pot1DBD (PDB ID: 4HIK). Pot1pC is colored green, 9mer is colored violet and atoms are colored by element. (B) Y224A mutation disrupts binding of Pot1pC to 9mer by nearly 10-fold. (C) The same Y224A mutation in Pot1-DBD (blue) has no effect on affinity relative to wild-type Pot1-DBD (red). (D) However, Y224A mutation apparently switches to the 12mer-binding mode based on the supershift (red arrow) at high protein concentration. G13C substitution also creates a supershift indicative of the 12mer-binding mode.
