Figure 1.
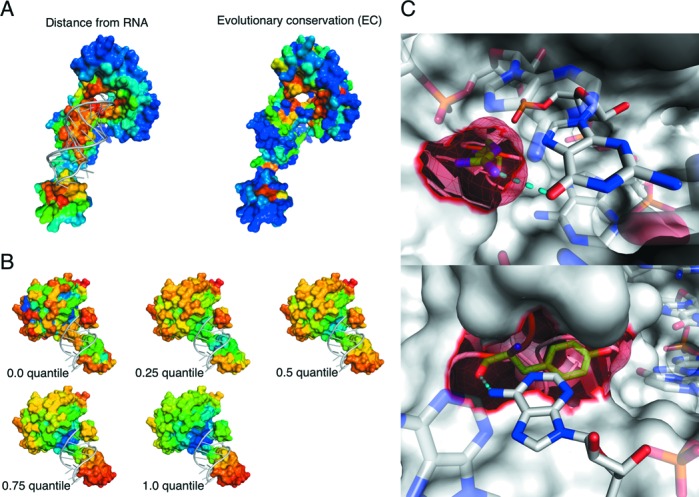
Novel features used in this work. (A) EC. A surface representation of the class-I A. fulgidus CCA-adding enzyme bound to a tRNA fragment (PDB ID: 3OVB). A distance map between protein and bound RNA with near (far) residues colored red (blue) is shown on the left. The EC value with high (low) colored red (blue) is shown on the right. (B) LN under a series of scales. LN values increase from blue to red. At each granularity level, warmer colors indicate convex residues, while cooler color represents concave residues. (C) Solvent ASA. A surface representation of RNase Cas6 (PDB ID: 4ILL) is shown. The protein makes both side-chain and backbone contacts with substrate RNA. Target residues (meshed) and nucleotides are represented by opaque sticks, connected by hydrogen bonds (dashed lines). The side chain of R268 protrudes and binds G15 (top). The backbone of Y168, which is mostly buried and forms part of a cleft, interacts with A5 (bottom). All figures of 3D structure representation in this work were generated by PyMOL Molecular Graphics System, Version 1.5, Schrödinger, LLC.
