Figure 1.
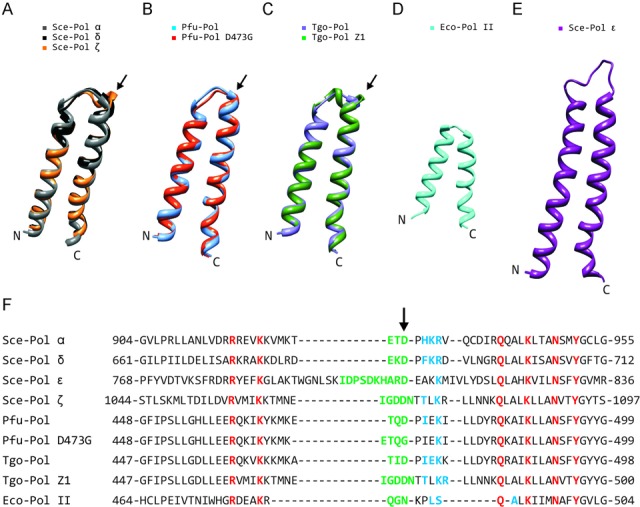
Comparison of fingers domains from family-B DNA polymerases. (A)–(C) Structural superimposition of the fingers regions from: (A) Saccharomyces cerevisiae Sce-Pol α (gray: pdb, 4FVM), Sce-Pol δ (black: pdb, 3IAY) and Sce-Pol ζ (orange: ‘Phyre2’ model); (B) Pyrococcus furiosus Pfu-Pol (blue: pdb, 2JGU) and Pfu-Pol (D473G) (red: ‘Phyre2’ model); (C) Thermococcus gorgonarius Tgo-Pol (purple: pdb, 1Tgo) and Tgo-Pol Z1 (green: ‘Phyre2’ model). With Sce-Pols α and δ, this region is virtually superimposable but the model of Sce-Pol ζ clearly shows an extended loop (arrowed). Similarly, the models of Pfu-Pol D473G and Tgo-Pol Z1 also indicate a slightly longer loop (arrowed) as compared to the wild types. (D) Fingers region of Eco-Pol II (cyan: pdb, 3K5O). (E) Fingers region of Sce-Pol ϵ (purple: pdb, 4M8O). (F) Amino acid alignments of the fingers domains. Amino acids in the loop region, based on the above structures/modelling, are coloured green. The arrow indicates the critical aspartic acid, which hydrogen bonds with amino acids (shown in blue) in the subsequent α-helix. The residues shown in red are very highly conserved and interact with incoming dNTPs. Sce: Saccharomyces cerevisiae; Pfu: Pyrococcus furiosus; Tgo: Thermococcus gorgonarius; Eco: Escherichia coli.
