Figure 3.
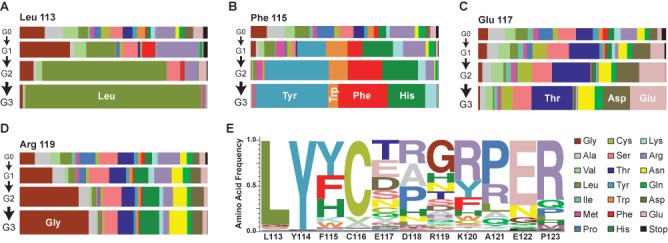
Functional determinants for the Leu113-Pro123 loop revealed by Sat-Sel-Seq. (A) Horizontal bar plots depicting the prevalence of amino acids from the four generational libraries, G0–G3. At position Leu113, stringent selection occurs in favor of the wild type (WT) residue. Alternative patterns of selection are evident at (B) position Phe115, where aromatic residues are selected by G3, (C) position Glu117 that remains highly diversified after selection or (D) position Arg119, where mutant variant R119G outcompetes the WT residue over generations. The individual data from other positions are provided in Supplementary Figure S4A. (E) Normalized logo plot for residues 113–123 summarizing the frequency of each amino acid at each position in the final selected library. An alternative representative of the G3 data scaled relative to the frequency of each amino acid in the G0 population is provided in Supplementary Figure S4B.
