Figure 1.
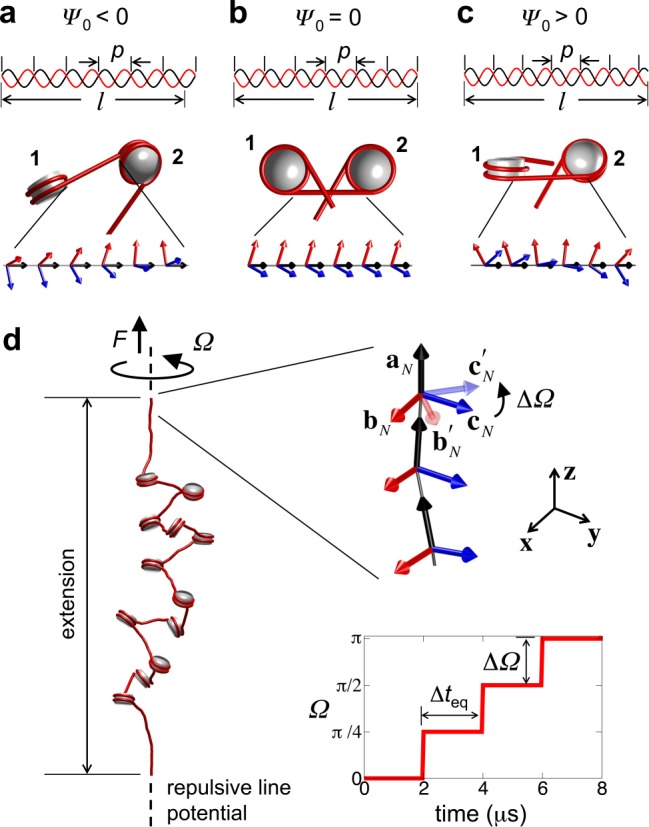
Rotational phase angle of nucleosomes and simulation setup. (a-c) Length and helical pitch of linker DNAs determines nucleosome phasing, and affects nucleosome topology: (b) l < nbp (where nb is an integer, equal to 6 in this study) leads to counter-clockwise rotation of downstream nucleosome 2 with respect to upstream nucleosome 1 (Ψ0 < 0) leading to more open linker DNAs; (b) l = nbp leads to in-phase nucleosomes (Ψ0 = 0); and (c) l > nbp leads to clockwise rotation of 2 with respect to 1 (Ψ0 > 0) leading to more crossed linker DNAs. (d) Simulation setup for studying the torsional response of nucleosome arrays. External twisting Ω is applied by rotating the last linker DNA frame by angle ΔΩ in a step-wise manner every Δteq while being subjected to a stretching force F. The repulsive line potential is drawn as dotted line and the DNAs and histone core are shown in red and gray, respectively.
