Figure 1.
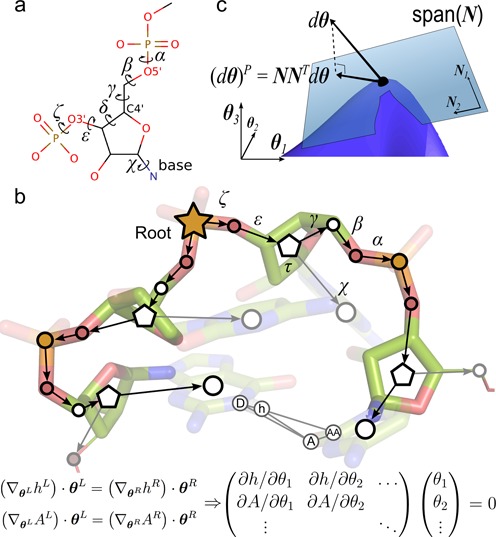
Kinematic representation of RNA. (a) A single nucleotide of RNA with its torsional degrees-of-freedom. (b) Edges in the directed spanning tree encode n torsional degrees-of-freedom 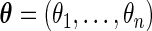 and vertices (circles) encode rigid bodies. Pentagons represent riboses, which have an additional internal degree-of-freedom governing their conformation (puckering). The hydrogen bond h-A closes a kinematic cycle, and is one of m distance constraints. As the position of the hydrogen atom h changes through perturbation of dihedral angles in the left branch of the tree, the new position of h should be matched by appropriate changes in the right branch, i.e.
and vertices (circles) encode rigid bodies. Pentagons represent riboses, which have an additional internal degree-of-freedom governing their conformation (puckering). The hydrogen bond h-A closes a kinematic cycle, and is one of m distance constraints. As the position of the hydrogen atom h changes through perturbation of dihedral angles in the left branch of the tree, the new position of h should be matched by appropriate changes in the right branch, i.e. 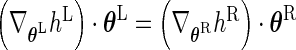 . Similarly, a change in position of heavy atom A from the right tree should be matched by changes in the left tree. These instantaneous distance constraints define the 6m × n Jacobian matrix
. Similarly, a change in position of heavy atom A from the right tree should be matched by changes in the left tree. These instantaneous distance constraints define the 6m × n Jacobian matrix 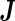 . (c) A schematic representation of the subspace of conformational space defined by the closure constraints. The subspace (blue surface) is highly nonlinear, but can be locally approximated by its tangent space, the null-space of
. (c) A schematic representation of the subspace of conformational space defined by the closure constraints. The subspace (blue surface) is highly nonlinear, but can be locally approximated by its tangent space, the null-space of 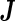 (translucent blue plane).
(translucent blue plane).
