Figure 2.
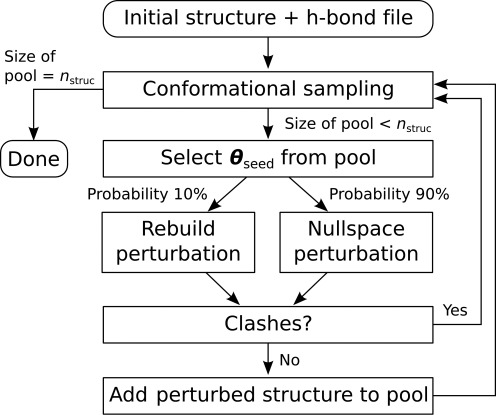
A flowchart of the KGSrna sampling algorithm. KGSrna takes as inputs an initial conformation and a file of hydrogen bonds A(N3)–U(H3) and G(H1)–C(N3) as distance constraints. Next, a pool of conformations is initialized with the input structure and then grown by repeatedly perturbing a randomly selected conformation from the pool with a rebuild or null-space perturbation at a 10/90 rate. If no clashes between atoms were introduced in the perturbed conformation, it is added to the pool. The procedure is repeated until a desired number nstruct of conformations is obtained.
