Figure 5.
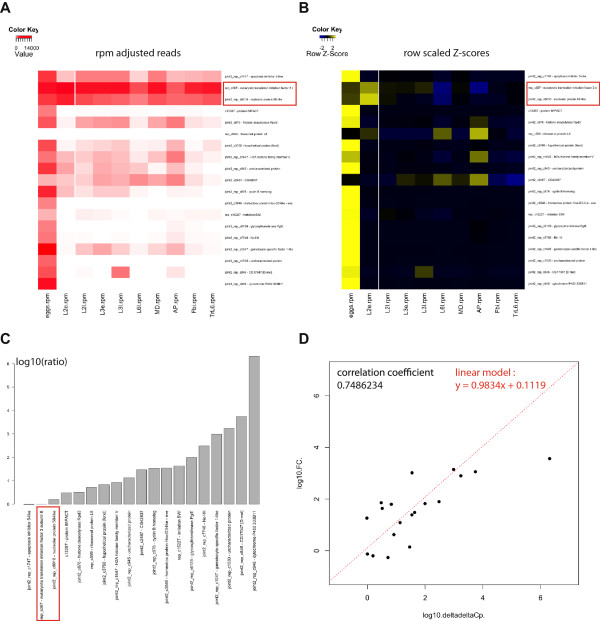
qPCR validation of differential expression. A. Heatmap representing the expression in rpm of each candidate transcript tested by qPCR. Genes are ordered from top to bottom from a lesser ratio between eggs and L2e samples, as measured in qPCR to a higher ratio. The red box shows the 2 genes that we used to normalize the qPCR. B. Same heatmap as in A. but showing expression as z-scores scaled by row to highlight the differential expression between eggs and L2e. C. Barplot showing the ratio measured in qPCR, using elongation factor 3 as a negative control for normalization. D. Scatter plot showing the correlation between fold changes measured by RNAseq (y axis) and ratios measured by qPCR for the tested genes. The two measurements have a correlation coefficient of 0.74. A linear regression model has been applied and is also shown on the same graph.
