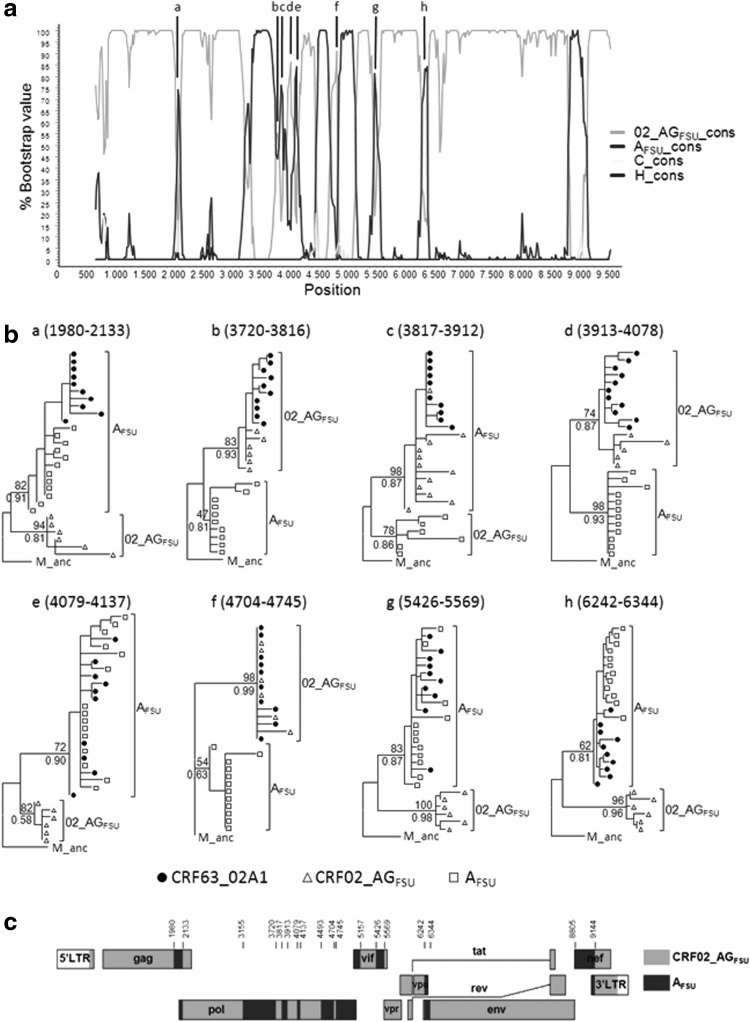
FIG. 2.
Analysis of the mosaic structure of CRF63_02A1. (a) Bootscan analysis of the consensus CRF63_02A1 near full-length genome sequence. Consensus AFSU and CRF02_AGFSU sequences were used as references and subtypes C and H as outgroups. The horizontal axis corresponds to positions in the HXB2 proviral genome sequence. (b) Phylogenetic trees of eight short (<200 nt) putatively recombinant segments of CRF63_02A1, as identified in the SimPlot bootscan analysis [signaled in (a)]; nucleotide positions in the HXB2 genome are indicated on top of each tree in parentheses; trees are rooted with the inferred group M ancestral sequence; branches corresponding to CRF63_02A1 viruses are marked with filled circles, those of CRF02_AGFSU viruses with triangles, and those of AFSU viruses with squares; bootstrap support, as determined with RAxML, and aLRT-SH-like support, as determined with PhyML, of AFSU and CRF02_AGFSU clades are shown on the left of the corresponding nodes, above and below, respectively, of the subtending branch. (c) Mosaic structure of CRF63_02AG inferred from SimPlot analysis and ML trees of partial segments. The HXB2 positions of breakpoints delimiting CRF02_AGFSU-derived and AFSU-derived segments are indicated on the top.