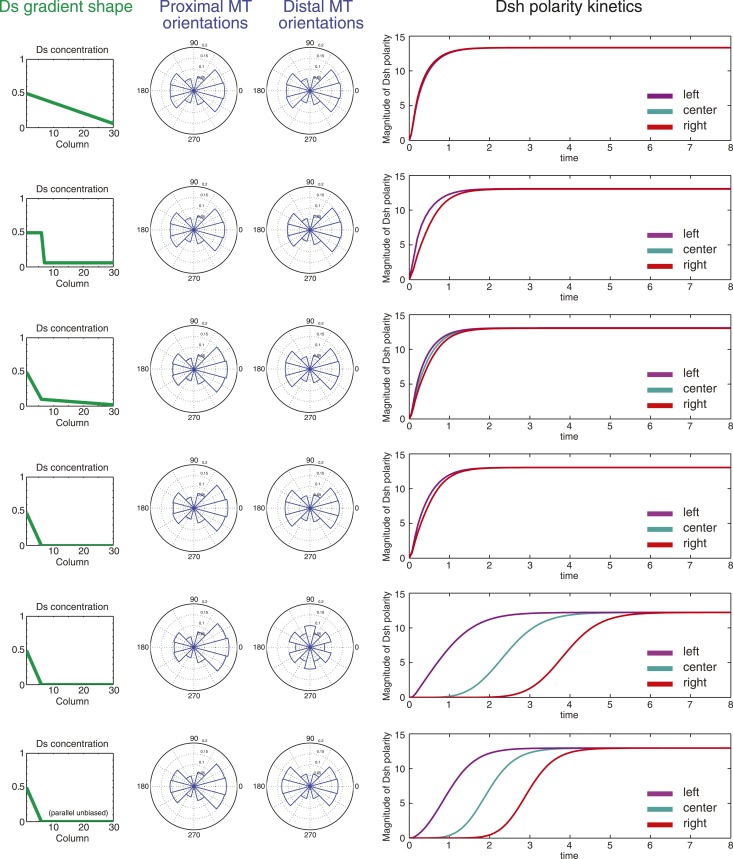
Figure 5. Simulations of polarization kinetics with different shapes of Ds gradients or imposed MT structures, using a mathematical model incorporating a simple representation of the Ft/Ds/Fj system to polarize MTs.
Input Ds gradients are shown on the left. Resulting (or imposed, on the right side of the last example) MT organization on the proximal or distal portion of the gradient are plotted (center). Kinetics of polarization of cells in column 8 (proximal = left), column 15 (center) or column 23 (distal = right; see Figure 5—figure supplement 1).