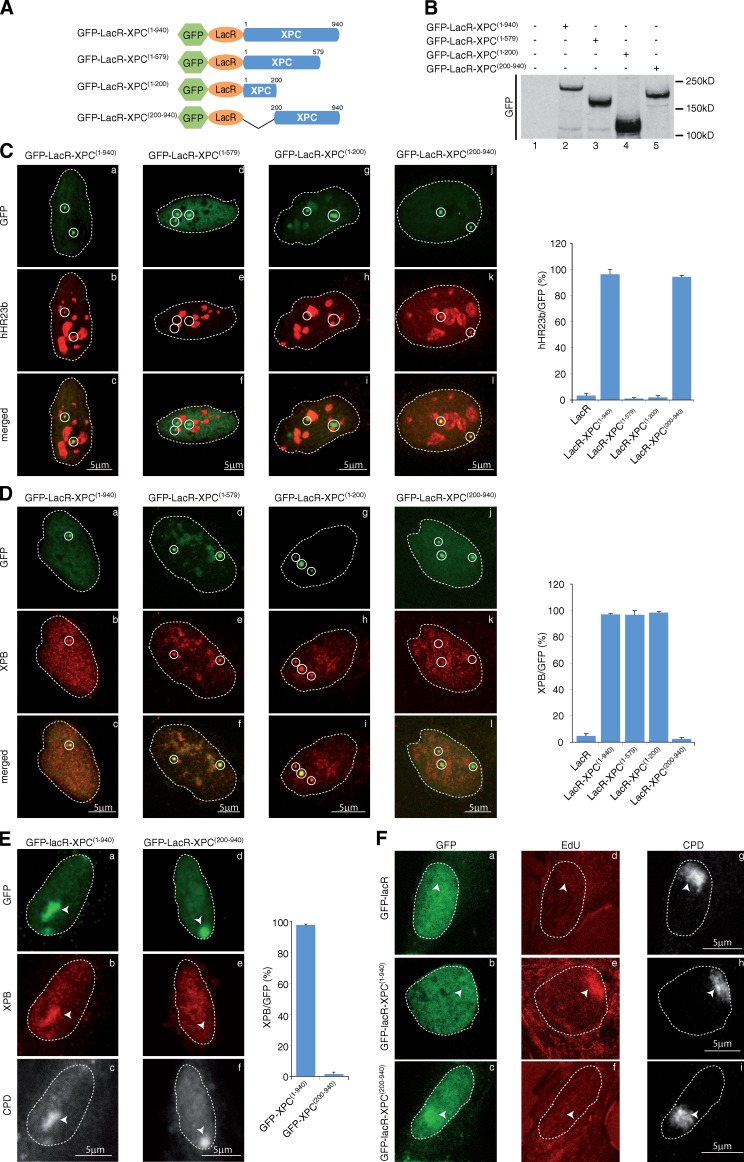
Figure 2.
Defining TFIIH- and hHR23b-interacting domains in XPC. (A) Schematic representation of the wild-type and mutant XPC constructs. For clarity, the sizes of the GFP (238 aa) and LacR (367 aa) were omitted. (B) Proteins from whole cell extracts (15 µg) of U2OS17-expressing wild-type or mutant XPC proteins were resolved by SDS-PAGE followed by Western blotting using anti-GFP antibody. (C and D) Recruitment of hHR23b (C) and XPB (D) to tethered GFP-LacR-XPC(1–940) (a–c), GFP-LacR-XPC(1–579) (d–f), GFP-LacR-XPC(1–200) (g–i), or GFP-LacR-XPC(200−940) (j–l). The values on the graphs represent the percentage of colocalization of each NER factor with GFP on the array based on three independent experiments with SD. Circles indicate LacO arrays. (E) After transfection of GFP-LacR-XPC(1–940) or GFP-LacR-XPC(200–940), XP-C cells were locally UV irradiated (150 J/m2), fixed 15 min later, and stained with antibodies raised against GFP, XPB, and UV-induced CPD. The values on the graphs represent the percentage of colocalization of XPB with GFP on the local UV array based on three independent experiments with SD. Arrowheads indicate locally irradiated areas. (F) After transfection of GFP-LacR, GFP-LacR-XPC(1–940), or GFP-LacR-XPC(200–940) (GFP), XP-C cells were locally UV irradiated (150 J/m2) and repair synthesis was analyzed by EdU incorporation at DNA damage (CPD). Arrowheads indicate locally irradiated areas.