Figure 3. Sir3 binds at two monomers per nucleosome and stabilizes the rod-like structure of chromatin.
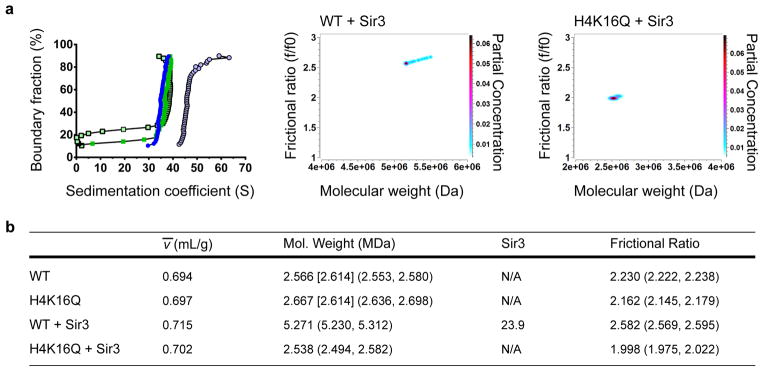
(a) Left panel, vHW of WT and H4-K16Q 12mer arrays +/− 2 Sir3 monomers per nucleosome. WT array is represented by dark blue closed circles, H4-K16Q by green closed squares, WT + Sir3 by light blue open circles, and H4-K16Q + Sir3 by light green open squares. Middle and right panels, GA-MC plots showing f/f0 vs. molecular weight for WT and H4-K16Q with Sir3. (b) 2DSA/GA-MC data of samples in (a) using experimentally-determined ν̄ values (see Supplementary Fig. 7 for examples). Numbers in brackets represent the expected molecular weight of a 601-177-12 array, and numbers in parentheses are 95% confidence intervals. Sir3 stoichiometry was calculated by subtracting the molecular weight of the array from the molecular weight of the array containing Sir3, divided by the molecular weight of 3xFLAG-tagged Sir3 (113 kDa).
