SUMMARY
Histone acetylation plays important roles in gene regulation, DNA replication, and the response to DNA damage, and it is frequently deregulated in tumors. We postulated that tumor cell histone acetylation levels are determined in part by changes in acetyl-CoA availability mediated by oncogenic metabolic reprogramming. Here, we demonstrate that acetyl-CoA is dynamically regulated by glucose availability in cancer cells and that the ratio of acetyl-CoA: coenzyme A within the nucleus modulates global histone acetylation levels. In vivo, expression of oncogenic Kras or Akt stimulates histone acetylation changes that precede tumor development. Furthermore, we show that Akt's effects on histone acetylation are mediated through the metabolic enzyme ATP-citrate lyase (ACLY), and that pAkt(Ser473) levels correlate significantly with histone acetylation marks in human gliomas and prostate tumors. The data implicate acetyl-CoA metabolism as a key determinant of histone acetylation levels in cancer cells.
INTRODUCTION
Rewiring of cellular metabolism in cancer cells is crucial for increased macromolecular biosynthesis, growth, and proliferation, and metabolic deregulation is now considered a hallmark feature of cancer cells (Hanahan and Weinberg, 2011; Ward and Thompson, 2012). In addition to directly supporting energetics and biosynthesis, a growing body of evidence suggests that metabolic enzymes may also promote tumorigenesis through functions that are not overtly metabolic. For example, the M2 isoform of pyruvate kinase, a glycolytic enzyme, has been reported to enter the nucleus and serve as transcriptional coregulator (Luo and Semenza, 2012). ATP-citrate lyase (ACLY), a lipogenic enzyme, is also present in the nucleus and plays a crucial role in regulating nuclear acetylation events, including histone acetylation (Wellen et al., 2009).
Metabolic changes in cancer cells are typically mediated by activation of oncogenes and/or loss of tumor suppressors. Hence, mutations that drive tumorigenesis also cause metabolic alterations. In addition to genetic mutations, epigenetic alterations play a key role in enabling malignant transformation and tumor growth (Azad et al., 2013; Shen and Laird, 2013), although mechanisms underlying epigenetic alterations in cancer are not fully clear. Notably, many chromatin-modifying enzymes depend on metabolic intermediates as cofactors or substrates and certain chromatin modifications have been shown to be sensitive to the cellular metabolic state [reviewed in (Kaelin and McKnight, 2013; Katada et al., 2012; Lu and Thompson, 2012; Yun et al., 2012)]. Links between cancer cell metabolism and DNA and histone methylation have been recently identified in tumors with specific mutations, such as those in isocitrate dehydrogenase (IDH1 and IDH2). Mutant IDH enzymes exhibit altered enzyme activity favoring conversion of α-ketoglutarate (αKG) into (R)-2-hydroxyglutarate, a metabolite that inhibits the activity of αKG-dependent JmjC-domain histone demethylases and TET enzymes, resulting in hypermethylation of DNA and histones [reviewed in (Losman and Kaelin, 2013; Ward and Thompson, 2012)]. Succinate dehydrogenase (SDH) mutations and nicotinamide N-methyltransferase (NNMT) overexpression also result in methylation changes in cancer cells (Killian et al., 2013; Letouze et al., 2013; Ulanovskaya et al., 2013; Xiao et al., 2012). In these specific cases, the epigenetic alterations are a direct result of altered activity of a particular metabolic enzyme. However, it is not known whether metabolism might more universally contribute to tumor epigenetic deregulation.
Histone acetylation is a dynamic chromatin mark, with important roles in gene regulation, DNA damage repair, and DNA replication. Global histone acetylation has also been implicated in contributing to pH balance and postulated to serve as a depot for acetate that could be mobilized as an energy source (Martinez-Pastor et al., 2013; McBrian et al., 2013). The metabolite acetyl-coenzyme A (acetyl-CoA) is the required donor substrate used by lysine acetyltransferase (KAT) enzymes. In cells cultured under standard conditions, glucose-derived carbon supplies the majority of acetyl-CoA used for histone acetylation, although glutamine and acetate also contribute (Evertts et al., 2013). In some conditions, fatty acid breakdown can also contribute to nuclear acetyl-CoA and histone acetylation (Donohoe et al., 2012). Acetyl-CoA is compartmentalized into mitochondrial and nuclear-cytoplasmic pools (Takahashi et al., 2006). Production of nuclear-cytoplasmic acetyl-CoA from glucose, glutamine, or fatty acids depends on export of citrate from the mitochondria, where it is cleaved by ACLY to generate acetyl-CoA and oxaloacetate. Acetate also supplies nuclear-cytoplasmic acetyl-CoA through acetyl-CoA synthetase (AceCS1/ ACSS2). Together, ACLY and AceCS1 are the primary enzymatic sources of acetyl-CoA outside of the mitochondria. Several studies have demonstrated that histone acetylation levels are highly sensitive to the availability of acetyl-CoA, in cell types from yeast to human (Cai et al., 2011; Donohoe et al., 2012; Friis et al., 2009; Takahashi et al., 2006; Wellen et al., 2009).
Given that production and utilization of metabolites is dramatically altered in cancer cells, and that histone acetylation is responsive to acetyl-CoA availability, we postulated that metabolic reprogramming in tumor cells might alter acetyl-CoA and histone acetylation levels in tumors. In this study, we provide evidence that metabolic reprogramming orchestrated by oncogenic activation of Akt drives changes in acetyl-CoA production and histone acetylation that are reflected in total histone acetylation levels in tumors. To our knowledge, this study is the first demonstration that metabolic reprogramming mediated by oncogenic activation of a signal transduction pathway plays a major role in regulation of the cancer cell epigenome.
RESULTS
Histone acetylation is variably sensitive to glucose in cancer cells
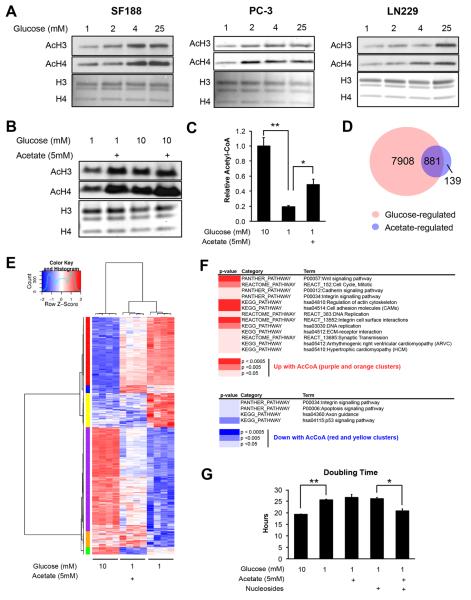
We previously showed that ACLY plays a key role in determining global histone acetylation levels in cancer cells. Histone acetylation is markedly reduced upon silencing of ACLY and can be restored by supplementing cells with the alternate acetyl-CoA source acetate, indicating that the ability of cells to produce acetyl-CoA can limit histone acetylation (Wellen et al., 2009). To determine whether glucose availability regulates histone acetylation levels in cancer cells, we examined a panel of cancer cell lines, including glioblastoma, prostate cancer, and breast cancer cell lines. Histone acetylation was sensitive to glucose availability in each of the cell lines examined, to varying degrees (Fig. 1A and S1A). As we have previously shown in other cell types (Wellen et al., 2009), silencing of ACLY inhibited glucose-dependent histone acetylation in LN229 cells (Fig. S1B).
Figure 1. Glucose availability regulates histone acetylation in several cancer cell lines.
(A) Acetylation of acid-extracted histones from cells cultured under indicated glucose conditions for 24 hours. Total histones were stained by Coomassie or Ponceau. (B) Acetylation of acid extracted histones from LN229 cells treated with 1 or 10 mM glucose, +/− 5 mM NaOAc for 24 hours (C) Relative levels of acetyl-CoA in LN229 cells after 24 hours of indicated treatment, mean +/− SEM of triplicates (*, p<0.05; **, p<0.005) (D) Venn diagram of genes regulated by glucose and/or acetate in LN229 cells, with the overlap representing genes designated as acetyl-CoA-regulated (E) Heatmap of 881 genes represented in the overlap from the Venn diagram. See Table S3 for associated gene list and cluster ID. (F) DAVID functional annotation of pathways regulated by acetyl-CoA, using the gene list identified from the indicated clusters on the heatmap. (G) Doubling time for LN229 cells treated as indicated for 24 hours, mean +/− SEM of triplicates (*, p<0.05; **, p<0.005).
In LN229 glioblastoma cells, proliferation slowed when cultured in 1 mM glucose, but between 2 and 25 mM glucose doubling time was equal, indicating that histone acetylation patterns are regulated by glucose independently of cell proliferation (Fig. S1C). Cells remained viable in all conditions (Fig. S1C). As glucose was progressively limited, glutamine consumption increased (Fig. S1C). Although glutamine can be used to supply nuclear-cytoplasmic acetyl-CoA, LN229 cells failed to sustain high histone acetylation levels in low glucose despite increased consumption of glutamine (Fig. 1A).
We next sought to define which histone lysines are most sensitive to glucose availability. LN229 cells were incubated for 24 hours in 1 mM or 10 mM glucose and histone modifications analyzed by mass spectrometry. Higher acetylation in 10 mM than 1 mM glucose was observed at all H2A, H3, and H4 tail lysines analyzed (Fig. S1D and Table S1). To confirm that this is due to reduced glucose carbon incorporation into acetylated histones, this analysis was repeated in 1 mM or 10 mM [U-13C6]-glucose. After 24 hours, enrichment was reduced in low glucose for all lysines analyzed (Table S2). However, the effects of glucose limitation became more pronounced as more lysines were acetylated in cis. For example, the histone H4 tail can be acetylated at lysines 5, 8, 12, and 16. In 1 mM [U-13C6]-glucose, significantly more tetra-acetylated histone H4 (AcH4) tails were enriched at 0 or 1 acetyl group, while in 10 mM, 3 or 4 acetyl groups were more frequently enriched (Fig. S1E). Thus, these data indicate 1) that on all tail lysines examined, acetylation is sensitive to glucose availability and 2) that glucose limitation has a greater impact on histone tails with multiple lysines acetylated in cis.
Supplementation of acetate restored histone acetylation levels in low glucose conditions (Fig. 1B). Correspondingly, acetyl-CoA levels were reduced in low glucose conditions and significantly rescued by addition of acetate (Fig. 1C). To determine whether specific gene sets correlated with these effects on histone acetylation, we performed RNA-sequencing to identify acetyl-CoA regulated genes (genes suppressed or induced by low glucose and reversed by acetate). Approximately 10% of glucose-regulated genes were also significantly regulated by acetate (Fig. 1D). Clustering analysis revealed that acetate regulated most genes in the same direction as glucose, suggesting that acetyl-CoA is a key mediator of glucose-dependent gene expression (Fig. 1E). Expression was confirmed for select acetyl-CoA-regulated genes by QPCR (Fig. S2A), and ChIP analyses at these genes showed that promoter histone acetylation correlated with expression (Fig. S2B). Analysis of curated pathways revealed enrichment for genes involved in cell cycle and DNA replication among acetyl-CoA-induced genes, suggesting that acetyl-CoA stimulates a pro-proliferative gene expression profile (Fig. 1F). Analogously, prior studies in yeast showed that acetyl-CoA promotes cell growth and division by promoting expression of genes involved in these processes (Cai et al., 2011; Shi and Tu, 2013). Although acetate supplementation was not sufficient to restore normal doubling time in low glucose conditions in LN229 cells (Fig. 1G), we considered that additional glucose-derived components might be required. Indeed, supplementation of acetate together with nucleosides (synthesis of which requires ribose generated in the pentose phosphate pathway), rescued doubling time, while neither component alone did so (Fig. 1G). These data suggest that acetyl-CoA facilitates expression of proliferation-related genes, as part of an integrated growth response.
Acetyl-CoA and Coenzyme A are key determinants of histone acetylation in cancer cells
If acetyl-CoA serves as a signal of nutrient sufficiency to promote growth and proliferation, cells must have mechanisms to detect changes in acetyl-CoA abundance. Work in yeast has shown that Gcn5 is crucial for mediating histone acetylation in response to acetyl-CoA availability (Cai et al., 2011; Friis et al., 2009). In yeast, acetyl-CoA concentrations oscillate within a range that could plausibly regulate yeast Gcn5, which has a Kd of 8.5 μM (Cai et al., 2011). However, human GCN5 binds acetyl-CoA much more tightly (Kd of 0.56 μM) (Langer et al., 2002). This suggests either that levels of acetyl-CoA are lower in human cells than in yeast or that the absolute concentration of acetyl-CoA is not the key regulatory factor for overall histone acetylation levels in human cells. Of note, many HATs including GCN5 are inhibited by the product coenzyme A (CoA) and bind acetyl-CoA and CoA with similar affinities. Hence, it has been postulated that the ratios of these metabolites might be the critical determinant for overall histone acetylation levels (Albaugh et al., 2011).
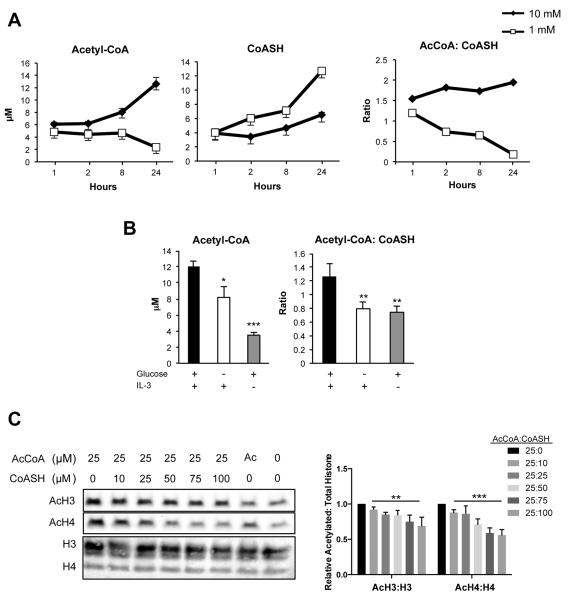
We therefore measured absolute acetyl-CoA and CoASH (reduced CoA) concentrations, normalizing to cell volume, in LN229 cells cultured in 1 or 10 mM glucose. Acetyl-CoA was measured at 6–13 μM in high glucose conditions and fell to 2–3 μM in low glucose. CoASH tended to rise in low glucose, and the ratio of acetyl-CoA: CoASH was found to be ~1.5– 2 throughout the time course in high glucose conditions and to fall below 1 under low glucose (Fig. 2A). We also examined acetyl-CoA and CoASH levels in a second cell line, IL-3-dependent hematopoietic cells, since the volume of these cells under standard conditions and during nutrient and growth factor withdrawal has been extensively characterized (Lum et al., 2005; Wellen et al., 2010). A similar range of acetyl-CoA concentrations (~3–12 μM) were measured in these cells as in LN229 cells, and both acetyl-CoA and acetyl-CoA: CoASH were significantly reduced when cells were deprived of either glucose or growth factor (Fig. 2B). The range of acetyl-CoA concentrations observed in each of these cell lines is comparable to that reported over the yeast metabolic cycle (Cai et al., 2011).
Figure 2. Acetyl-CoA: CoASH ratio is a determinant of histone acetylation in cancer cells.
(A) Molar concentrations of acetyl-CoA, CoASH, and ratio of acetyl-CoA: CoASH over 24 hours in LN229 cells, mean +/− SEM of triplicates. Result is representative of 3 independent experiments. (B) IL-3-dependent bax−/−bak−/− cells were cultured for 48 hours +/− glucose, +/− IL-3 as indicated. Acetyl-CoA and CoASH were measured and normalized to cell volume, mean +/− SD of triplicates. Significance as compared to Glc+IL-3+ samples (**, p<0.005, ***, p<0.0005) (C) Representative Western blot of acetylated histones upon incubation of isolated nuclei with varying concentrations of acetyl-CoA and CoASH. Total histones were stained by Ponceau. Data was quantified from four independent experiments with 25:0 value set to 1, mean +/− SEM. Repeated measures one-way ANOVA with post test for linear trend was performed for 25:10–25:100 values. Post test for linear trend significance for AcH4: H4, p<0.0001 (***); for AcH3: H3, p=0.0034 (**).
To specifically test whether histone acetylation levels respond to both acetyl-CoA and CoA levels, nuclei were isolated from LN229 cells and incubated with defined concentrations of acetyl-CoA and CoASH, which can freely enter the nucleus through the nuclear pore complex. We observed that in the presence of high acetyl-CoA, addition of CoASH suppressed histone acetylation in a dose-dependent manner (Fig. 2C). Together, these data indicate that levels of both acetyl-CoA and CoA in the nucleus can impact histone acetylation levels. Moreover, the ratio of acetyl-CoA: CoASH is glucose-sensitive and tracks with total acetyl-CoA.
Expression of KrasG12D in the mouse pancreas promotes increases histone acetylation prior to tumor development
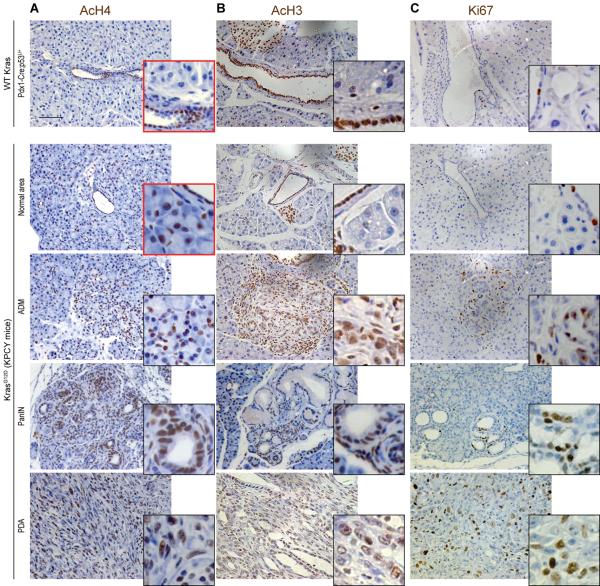
Since histone acetylation is sensitive to metabolic state and metabolism is altered in cancer cells, we next asked whether oncogene activation could drive changes in histone acetylation in vivo. Oncogenic Kras mutations are found in the vast majority of human pancreatic adenocarcinoma (PDA) cases, and recent studies have demonstrated that mutant Kras in pancreatic cancer cells drives extensive metabolic rewiring (Son et al., 2013; Ying et al., 2012). We observed that in the pancreas of healthy wt mice, acinar cells exhibited very low levels of AcH3 and AcH4, as detected by immunohistochemistry (IHC), although ductal cells were strongly positive for these marks (Fig. 3A). In contrast, in mice expressing oncogenic Kras [LSL-KrasG12D; p53L/+; Pdx1-Cre (KPC) mice], areas of acinar-to-ductal metaplasia (ADM), precancerous pancreatic intraepithelial neoplasia (PanIN), and PDA exhibited high levels of AcH3 and AcH4 (Fig. 3, A and B). PDA may arise from acinar cells (Morris et al., 2010), and strikingly, elevated AcH4 was observed in acinar cells even prior to increases in proliferation, as assessed by Ki67 immunostaining (Fig. 3C), or the appearance of any histological abnormalities (Fig. 3A and S3). Thus histone acetylation levels are elevated upon KrasG12D expression in mouse pancreas early in the process of tumor development.
Figure 3. Oncogenic Kras increases histone acetylation in vivo.
Pancreata from mice expressing Kras-G12D (KPCY, previously described(Rhim et al., 2012)) were harvested at either 6 weeks (KPCY-Normal Area), 8 weeks (KPCY-PanIN) or 10 weeks (KPCY-PDA) of age, along with pancreata from control (Kras WT) mice (n≥3, each group). Immunohistochemistry against AcH4 (A), AcH3 (B) or Ki67 (C) was performed on paraffin-embedded tissue sections and nuclei were counterstained with hematoxylin. Representative images are shown, with magnification of areas of interest. Scale bar: 50 μm. See Fig. S3 for quantification of AcH4 in acinar cells in Kras WT and KPCY mice.
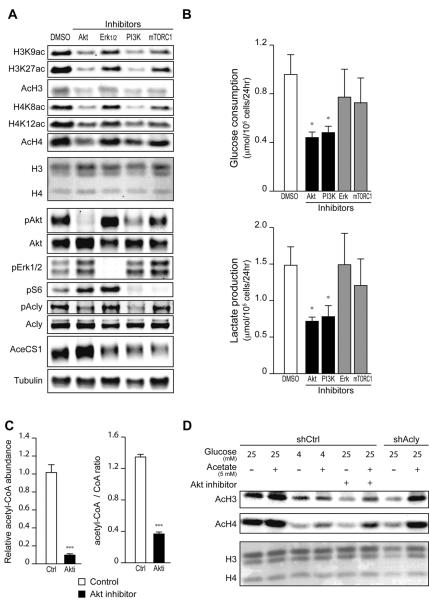
We next interrogated signaling pathways downstream of Kras to identify those responsible for elevating histone acetylation in primary murine PanIN cells. Reductions in histone acetylation were observed with each of the inhibitors that we tested (Fig. 4A). PI3K and Akt inhibition also significantly reduced glucose uptake and lactate consumption and suppressed ACLY phosphorylation (Fig. 4, A and B). Phosphorylation at Ser455 has been shown to increase ACLY's activity (Potapova et al., 2000). On the other hand, the level of AceCS1, which produces nuclear-cytoplasmic acetyl-CoA from acetate, was somewhat suppressed by PI3K, but not by Akt, inhibition in these cells (Fig. 4A). Acetyl-CoA and acetyl-CoA: CoASH levels were potently suppressed upon Akt inhibition (Fig. 4C), and histone acetylation levels in Akt inhibitor-treated cells increased upon supplementing cells with acetate (Fig. 4D). Suppression of histone acetylation by Akt inhibition was comparable to that observed upon either glucose limitation or ACL silencing, both of which were also rescued by acetate (Fig. 4D). In primary murine PDA cells, we similarly observed suppression of histone acetylation and glucose uptake by PI3K or Akt inhibition, as well as acetate rescue of histone acetylation in Akt inhibitor- or low glucose-treated cells (Fig. S4, B–D). These data indicate that in premalignant and cancerous cells expressing oncogenic Kras, histone acetylation is regulated in part by Akt-dependent regulation of acetyl-CoA metabolism.
Figure 4. Oncogenic Kras enhances histone acetylation in vitro in an Akt- and ACL-dependent manner.
(A) Mouse pancreatic primary cells derived from KPCY mouse PanIN lesions were treated with indicated inhibitors for 24 hours. Acetylation of histones and phosphorylation of signaling proteins were assessed by Western Blot in acid extracts and RIPA lysates, respectively. Ponceau staining is shown as loading control for histones. (B) Glucose consumption and Lactate production were measured in PanIN-derived primary mouse cells treated as in (A), mean +/− SD of triplicates (*, p<0.05). (C) Acetyl-CoA and CoASH levels were measured in PanIN cells, +/− Akt inhibitor, mean +/− SD of triplicates (***, p<0.001). (D) PanIN-derived primary cells were transduced with control (shCtrl) or ACL-targeting (shACL) short hairpin RNA. Cells were cultivated under indicated glucose concentrations, +/− Akt inhibitor, +/− 5 mM acetate for 24 hours. Histones were acid-extracted and analyzed by Western Blot. Ponceau staining is shown as loading control for histones.
Akt activation sustains histone acetylation in nutrient-limited conditions
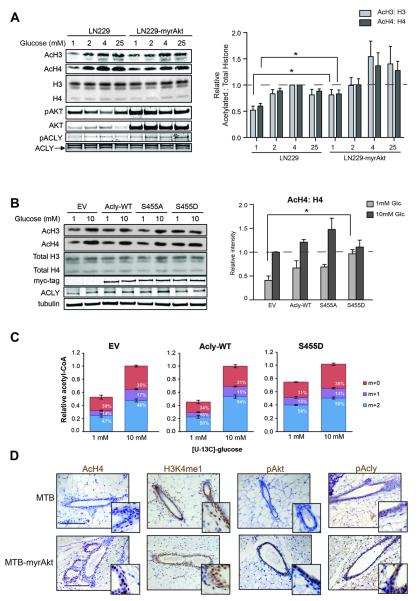
To probe further into the mechanism whereby Akt regulates histone acetylation, we examined the effects of constitutive Akt activation in cancer cells by comparing parental LN229 cells with LN229 cells in which a constitutively active form of Akt, myristoylated Akt (myrAkt), was stably expressed. LN229-myrAkt cells have been shown to consume more glucose but maintain a similar proliferation rate as parental cells (Elstrom et al., 2004). Levels of histone acetylation in high glucose conditions were not markedly different between the two cell lines. However, when cultured in low glucose, LN229-myrAkt cells sustained a significantly higher level of histone acetylation than parental cells (Fig. 5A). Similar results were also obtained in SF188 and SF188-myrAkt cells (Fig. S5A). Time course analysis indicated that myrAkt expression extended the timeframe over which cells could maintain histone acetylation levels in low glucose (Fig. S5B).
Figure 5. Akt activation allows sustained histone acetylation in glucose-limited conditions.
(A) Western blot analysis of proteins and histones from LN229 and LN229-myrAkt cells. Quantitation represents 5 independent experiments with LN229 4mM samples set to 1, mean +/− SEM (*, p ≤0.05). (B) Western blot analysis of histone acid extracts from LN229 cells stably expressing vector (EV), wt ACL, ACL-S455A, or ACL-S455D. Cells were treated with 1 or 10mM glucose for 24 hours before lysis. Quantitation represents the ratio of acetylated to total histones in four independent experiments with EV 10 mM set to 1, mean +/− SEM (*, p≤0.05) (C) Relative acetyl-CoA concentrations and percent enrichment after treatment in 1 mM or 10 mM [U-13C6]-glucose for 20 hours, mean +/− SEM of triplicates. Total acetyl-CoA levels as well as M+2 acetyl-CoA (enriched from glucose) were significantly suppressed (p≤0.05) in 1 mM as compared to 10 mM glucose in EV and WT-ACL cells, but not ACL-S455D cells. (D) Control (MTB) and transgenic (MTB-tAkt) mice were administered doxycyclin for 96 hours (n≥5, each group). Immunohistochemistry was performed on paraffin-embedded mammary tissue sections. Representative images are shown, along with magnification of areas of interest. Scale bar: 50 μm.
Glucose deprivation results in depletion of ACLY's substrate citrate in LN229 cells (Fig. S6A). In myrAkt-expressing cells, citrate levels were lower than in the parental cells, even in high glucose conditions, and loss of citrate was accelerated upon glucose deprivation (Fig. S6A). In vitro studies of ACLY enzymatic activity have demonstrated that phosphorylation at Ser455 increases the enzymatic activity of ACLY, resulting in a 6-fold increase in V(max) (Potapova et al., 2000). Hence, increased phosphorylation of ACLY by Akt could potentially enable sustained acetyl-CoA production and thereby histone acetylation even if availability of the ACLY substrate citrate is reduced. To test whether ACLY phosphorylation is sufficient to sustain high levels of histone acetylation in low glucose, we expressed wt ACLY and ACLY Ser455 phospho-mimetic (S455D) and phospho-mutant (S455A) proteins in LN229 cells. Expression of ACLY-S455D enabled high levels of histone acetylation to be sustained in low glucose, similar to that observed with myrAkt expression (Fig. 5B). A trend towards higher histone acetylation in low glucose was also noted with expression of wt ACLY. These results suggest that ACLY is a key downstream effector of Akt in promoting histone acetylation, particularly when nutrients are limited.
This result raised the question of where cells obtain the carbon for histone acetylation in cells with constitutive Akt/ACLY activation but limited glucose. Recent studies have shown that under certain conditions such as hypoxia, glutamine can be reductively carboxylated to generate citrate and supply lipogenic acetyl-CoA and that depletion of citrate or an elevated α-ketoglutarate: citrate ratio is necessary for this effect (Fendt et al., 2013; Gameiro et al., 2013). We hypothesized that citrate depletion observed with myrAkt expression might stimulate glutamine reductive carboxylation. However, analysis of citrate isotopologues following exposure to [13C5 15N2]-glutamine revealed that glutamine continues to be oxidized in myrAkt-expressing cells and that little to no reductive carboxylation occurred in either high or low glucose conditions (Fig. S6B). On the other hand, acetyl-CoA, though significantly depleted in control or wt ACL-expressing cells in low glucose, retained a comparable percent enrichment from glucose (M+2 acetyl-CoA) when cultured in either 1 mM or 10 mM [U-13C6]-glucose (Fig. 5C). Moreover, in ACLY-S455D-expressing cells, both total and M+2 acetyl-CoA resisted depletion in low glucose (Fig. 5C), suggesting that even when glucose is limited, it remains a major source of acetyl-CoA in this context.
Hence, the data indicate that Akt promotes acetyl-CoA production and histone acetylation through combined effects on 1) promoting the uptake and metabolism of glucose and 2) promoting phosphorylation and activation of ACLY to facilitate continued acetyl-CoA production even when its substrate citrate is limited.
Akt activation acutely promotes histone acetylation in vivo
To directly test whether acute activation of Akt regulates histone acetylation in vivo, we exploited a doxycycline-inducible mouse model of breast cancer, in which myrAkt is activated in mammary epithelial cells upon administration of doxycycline to mice through the drinking water (Gunther et al., 2002). pAkt and pACLY levels increased with myrAkt expression, as expected (Fig. 5D). In control animals, relatively weak staining for AcH4 was detected in ductal epithelial cells. Remarkably, however, upon induction of Akt, pronounced increases in histone acetylation were detected within 96 hours of doxycycline administration (Fig. 5D). On the other hand, another activating mark, H3K4me1, was not obviously regulated, suggesting that histone acetylation - not histone marks in general- may be specifically impacted by Akt activation. Hence, acute activation of Akt in vivo increases global histone acetylation levels.
These data indicate that activation of Akt acutely promotes histone acetylation. We next investigated whether Akt activation is associated with histone acetylation in established human tumors.
Histone acetylation levels correlate with pAkt in human glioma
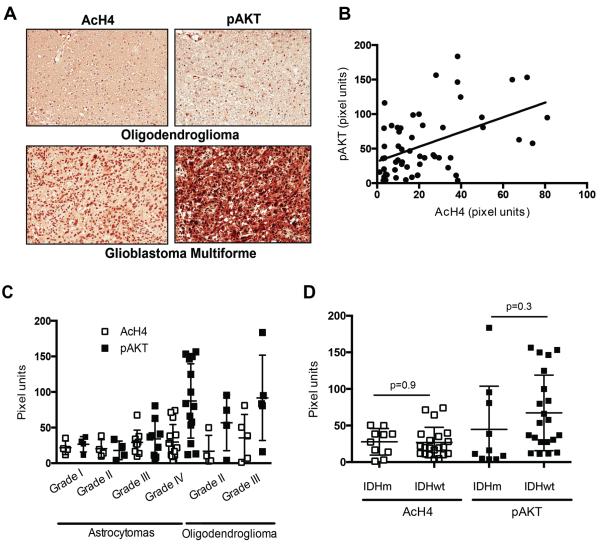
The PI3K/Akt pathway is frequently activated in GBM as a result of mutations resulting in activation of EGFR or PI3K or loss of PTEN (Lino and Merlo, 2011). We examined a diverse panel of human gliomas to determine whether AcH4 is associated with pAkt levels. A total of 56 samples on a tumor tissue microarray (TMA) were analyzed for pAkt and AcH4 (Fig. 6A). Each tumor sample was scored in a blinded manner by a neuropathologist and given a combined score for percent of positive cells and intensity of staining (H-score) for both pAkt and AcH4. A statistically significant positive correlation was found between pAkt and AcH4, indicating that Akt activation is associated with increased histone acetylation in human tumors (Fig. 6B). pAkt levels tended to increase with tumor grade, particularly in astrocytomas, although no significant relationship was observed between tumor grade and AcH4 (Fig. 6C). The relationship between pAkt and AcH4 is therefore not secondary to effects of tumor grade. IDH mutations occur frequently in gliomas and alter both cell metabolism and epigenetics (Losman and Kaelin, 2013). Of particular relevance, IDH mutations cause hypermethylation of histones (Lu et al., 2012), suggesting the possibility that histone acetylation might be altered secondarily to methylation effects in IDH mutant tumors. H3K9me3 was previously shown to be elevated in IDH mutant tumors in this patient set (Venneti et al., 2013a), and we found no significant relationship between H3K9me3 and pAkt levels (not shown). Moreover, AcH4 levels were not different between IDH wt and IDH mutant tumors (Fig. 6D).
Figure 6. pAkt correlates with histone acetylation in human glioma.
(A) Representative images (20X) of pAkt and AcH4 in a grade IV GBM and a grade II oligodendroglioma. (B) pAkt expression showed a significant correlation with AcH4 levels in 56 human glioma samples (r=0.4721, p=0.0002). (C) pAkt and AcH4 in astrocytic tumors (grade IV astrocytomas = GBM) and oligodendrogliomas, mean +/−SD. (D) pAkt and AcH4 levels in 10 IDH mutant and 21 IDH wt gliomas, mean +/− SD.
Histone acetylation levels correlate with pAkt in human prostate cancer
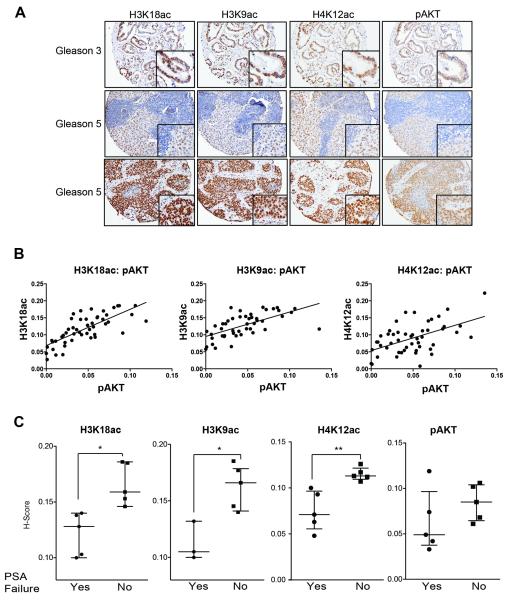
In human prostate tumors, significant correlations have been reported between levels of histone acetylation marks with tumor grade (H3K18ac and H4K12ac), as well as with the risk of tumor recurrence (H3K18ac, H3K9ac, H4K12ac) (Bianco-Miotto et al., 2010; Seligson et al., 2005). The PI3K-Akt pathway is frequently activated in prostate cancer due to PTEN loss or other mutations, and inhibition of Akt in the PTEN null prostate cancer cell lines PC-3 and C4–2 reduced overall levels of histone acetylation (Fig. S7A). We investigated levels of three histone acetylation marks- H3K18ac, H3K9ac, and H4K12ac- and their association with pAkt(Ser473) in a panel of human prostate tumors (Fig. 7A). The cohort consisted of patients with either metastatic or localized disease, with primary tumor grades ranging from 3–5 on the Gleason scale. Tumors were scored for both the percentage of positive cells and the intensity of staining. A striking positive correlation was observed between pAkt and each of the histone acetylation marks (Fig. 7B). Levels of each histone acetylation mark were also strongly correlated with one another in tumors (Fig. S7B). Percentages of nuclei positive for H3K18ac were similar between patients with metastatic as compared to localized disease, while H3K9ac and particularly H4K12ac scored positive in a higher percentage of nuclei in patients with metastatic disease (Fig S7C). H4K12ac was also higher in Gleason 4 and 5 tumors than Gleason 3 samples (Fig. S7D). Together the data from human prostate cancer and glioma suggest that Akt activation is a major determinant of histone acetylation levels in tumors.
Figure 7. pAkt correlates with histone acetylation levels in human prostate cancer.
(A) Representative images of H3K18ac, H3K9ac, H4K12ac, and pAkt expression detected by immunohistochemistry in Gleason grade 3–5, metastatic (n=25) and non-metastatic (n=24) prostate cancer tumors. (B) H-scores were determined and correlations between marks determined. pAkt expression showed a significant correlation with: H3K18ac levels (r=0.7452, p≤0.0001); H3K9Ac (r=0.6283, p≤0.0001); and H4K12ac (r=0.5276, p≤0.0001). (C) Box plot showing H-scores for H4K12ac, H3K18ac, H3K9ac, and pAkt in prostate cancer tumors in patients who developed PSA failure (Yes) or not (No) (**, p<0.005, *, p<0.05).
Histone acetylation as a predictive biomarker for therapeutic failure
For 10 of the prostate cancer patients with localized disease, biochemical failure (PSA recurrence) data was available. Strikingly, levels of histone acetylation were highly predictive of which patients would later exhibit biochemical failure. The five patients with the lowest levels of H4K12ac, H3K18ac, and H3K9ac developed biochemical failure, whereas the five with the highest levels of these marks did not (Fig. 7C). These findings agree with those of Seligson and colleagues, showing that low levels of histone modifications such as H3K18ac are associated with a poor prognosis in prostate cancer (Seligson et al., 2005). Within these 10 samples, the relationship between pAkt and each histone acetylation mark held (Fig. S7E), although pAkt levels did not predict biochemical failure in these 10 samples (Fig. 7C). Hence, histone acetylation levels might be a valuable biomarker to predict tumor relapse.
DISCUSSION
We report that oncogenic Akt activation is a key determinant of global histone acetylation levels in cancer cells. We provide evidence that this occurs through Akt-dependent metabolic reprogramming to promote high acetyl-CoA production. Akt ensures continuous acetyl-CoA production even during nutrient limitation by promoting the phosphorylation and activation of ACLY. To our knowledge, this is the first report that implicates metabolic reprogramming mediated by oncogenic activation of a signal transduction pathway as a major factor underlying tumor epigenomic regulation.
Elucidating the regulation of tumor histone acetylation levels is important clinically, and the mechanisms that control tumor histone acetylation are currently poorly understood. Our data suggest that metabolic effects contribute to determining histone acetylation levels in tumors. Other relevant factors in addition to acetyl-CoA metabolism include levels of KATs and HDACs, as well as tumor pH. The significance of global histone acetylation levels is not yet fully clear. Several studies have examined the relationship between histone acetylation levels and tumor recurrence and patient survival in various cancer types [reviewed in (Chervona and Costa, 2012; Kurdistani, 2007)]. Significant correlations have been shown in several studies, although high histone acetylation has been associated with both better and worse prognoses, likely reflecting differences in types of cancer, tumor grade, therapeutic approaches, and sample stratification. Our data suggest that low levels of histone acetylation may predict poor patient outcome in prostate cancer, similar to the findings of Seligson et al (Seligson et al., 2005). Further study is needed to understand how global histone acetylation levels impact tumor growth and progression, as well as treatment responses.
It will be crucial to further investigate the functions of metabolic control of acetylation, as well as the role of ACLY levels and phosphorylation, in regulating gene expression and promoting tumorigenesis. ACLY silencing is known to inhibit cancer cell proliferation and suppress tumor growth (Bauer et al., 2005; Hatzivassiliou et al., 2005; Migita et al., 2008). While suppression of de novo lipogenesis in the absence of ACLY is certainly a key component of this anti-tumor effect, a growing literature indicates that acetylation also plays a crucial role in promoting anabolic metabolism and growth. Acetyl-CoA has been shown to promote growth and proliferation in yeast, through histone acetylation at genes involved in these processes (Cai et al., 2011; Shi and Tu, 2013). Moreover, acetyl-CoA and acetylation regulate many metabolic enzymes (Guan and Xiong, 2011), and a role for acetyl-CoA in suppressing autophagy has also recently been uncovered (Eisenberg et al., 2014; Marino et al., 2014). We show here that many proliferation-related genes are acetyl-CoA-regulated in glioblastoma cells, correlating with overall histone acetylation levels, as well as histone acetylation at these genes. Thus, our study adds to a growing literature that implicates acetyl-CoA in providing pro-growth cues to the cell.
We have focused this study on the role of the PI3K/Akt pathway in regulation of acetyl-CoA production and histone acetylation. Many oncogenes and tumor suppressors are now recognized to regulate cellular metabolism. Microenvironmental conditions such as hypoxia also potently reprogram cellular metabolism. Moreover, in addition to acetyl-CoA, metabolites such as UDP-GlcNAc, S-adenosylmethionine (SAM), and α-ketoglutarate are also required by chromatin modifying enzymes and could potentially impact the epigenome if their levels are altered in tumors (Lu and Thompson, 2012). It is therefore conceivable that altered metabolite utilization substantially alters chromatin in many or most cancer cells.
Intense effort is currently aimed at targeting both cancer cell metabolism and cancer epigenetics (Dawson and Kouzarides, 2012; Vander Heiden, 2011). If metabolism is indeed a critical mediator of cancer cell epigenetic deregulation, it is possible that epigenetic alterations could be reversed through therapeutics directed at metabolic targets. Improved understanding of metabolic regulation of the epigenome could point towards contexts in which existing epigenetic therapies such as HDAC inhibitors would be most effective or could open doors to development of more specific therapeutics targeting the intersection of metabolism and epigenetics. The finding that the PI3K/Akt pathway is a key determinant of histone acetylation levels in tumors represents an important step forward in understanding the extent to which oncogenic metabolic reprogramming could control the epigenome.
EXPERIMENTAL PROCEDURES (Additional details provided in Supplement)
Reagents and Cell Lines
LN229-myrAkt cells have been previously described (Elstrom et al., 2004). LN229-myrAkt and SF188-myrAkt cells were a gift of C.B. Thompson, MSKCC. LN229-ACLY, -S455A, and – S455D and empty vector stable cell lines were generated by transfecting cells with pEF6, pEF6-ACLY(wt), pEF6-ACLY(S455A), or pEF6-ACLY(S455D) constructs and selecting. See Supplement for specific culture conditions and information on inhibitors and antibodies used.
YSI metabolite and doubling time measurements
Glucose and glutamine consumption and lactate production were measured using a YSI 7100 Bioanalyzer. Measurements were conducted over a 24 hour time period and normalized to cell number area under the curve, as previously described (Londono Gentile et al., 2013). Briefly, metabolite consumption was defined as v = V(xmedium control - xfinal)/A, where v is metabolite consumption/production, V is culture volume, x is metabolite concentration, and A is cell number area under the curve. A was calculated as N(T)d/ ln2(1–2-T/d), where N(T) is the final cell count, d is doubling time, and T is time of experiment. Doubling time was calculated as d = (T)[log(2)/log(Q2/Q1)], where Q1 is starting cell number and Q2 is final cell number, as determined by manual counting using a hemocytometer.
Mass spectrometry analysis of histones
For 12C study, LN229 cells in triplicate wells were incubated for 24 hours in 1 mM or 10 mM glucose. For 13C study, LN229 cells were first cultured in 1 mM glucose overnight. Triplicate wells of cells were then treated with 1 or 10 mM [U-13C]-glucose for 2 or 24 hours. Histones were acid extracted, as described in supplement. Total histones were subjected to chemical derivatization using propionic anhydride (Sigma-Aldrich) and digested with sequencing grade trypsin at a 10:1 substrate to enzyme ratio for 6 hours at 37°C. The digested peptides were treated with an additional round of propionylation for the purpose of adding propionyl group to the newly generated N-terminus. Peptides were desalted using C18 extracted mini disk (Empore 3M, MN, USA) and dissolved in 0.1% formic acid. Approximately 1ug of each sample was loaded via an autosampler (EASY-nLC, Thermo Fisher Scientific Inc) onto a homemade 75μm reversed phase analytical column packed with 15cm C18-AQ resin (3μm particle sizes, 120 Å pore size) at a rate of 550nl/min. Peptides were chromatographically resolved on a 66-min 2–98% solvent B gradient (solvent A = 0.1% formic acid, solvent B = 100% acetonitrile) at a flow rate of 250 nL/min. The eluted peptides were electrosprayed through a PicoTip emitter (New Objective Inc, Woburn, MA) into and detected by LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific Inc) with a resolution of 60,000 for full MS spectrum followed by MS/MS spectra obtained in the ion trap.
After the MS data were generated, an in-house program was developed to calculate the relative intensity of 13C enriched acetyl groups. For example, there are four acetyl groups in H4 4–17 4ac, with 2-Dalton mass shift between adjacent groups. Because the isotope patterns of adjacent groups overlap, the intensity from the former group should be subtracted. After calibration, the intensity of each group divided by the total intensity is the relative intensity.
RNA-sequencing
LN229 cells were incubated for 24 hours in 1 mM or 10 mM glucose, or 1 mM glucose + 5 mM sodium acetate. RNA was isolated using Trizol reagent and submitted to the Functional Genomics Core at the University of Pennsylvania for library preparation and sequencing. Details of data analysis and clustering may be found in the supplement. Data is deposited in GEO under accession number GSE57488.
LC-MS metabolite measurements
Acetyl-CoA and CoASH measurements were conducted as previously described (Basu and Blair, 2012; Basu et al., 2011). Detailed protocol is included in Supplement. For molar concentrations, measurements were normalized to cell volume, as determined using a Coulter Z2 particle counter, as previously described (Wellen et al., 2010). Protocol for citrate and glutamate analysis is also described in supplement.
Histone acetylation in isolated nuclei assay
Nuclei were isolated from approximately 9 × 106 adherent LN229 cells by using a cell lifter and cold NIB buffer (15mM Tris-HCL pH7.5, 60mM KCl, 15mM NaCl, 5mM MgCl2, 1mM CaCl2, 250mM sucrose, 0.1% NP-40). Nuclei were pelleted at 600 rcf for 5 min in 4°C and followed by two washes using NIB buffer without NP-40. Nuclei were suspended with indicated concentrations of acetyl-CoA and CoASH in 200 μL NIB buffer without NP-40 for 1 hour at 37°C. After incubation, nuclei were pelleted at 600 rcf for 5 min and subject to acid extraction (described in Supplement).
Immunohistochemistry and scoring of murine tissues
For histological evaluation, tissue samples were harvested as described (Gunther et al., 2002; Rhim et al., 2012). Immunohistochemistry was performed on paraffin-embedded sections. Tissue sections were dewaxed and rehydrated. Antigen retrieval was performed by boiling samples in Citrate Buffer pH6 for 20 min (for AcH4, AcH3 and H3K4me) or in Tris-EDTA 10mM Buffer pH9 for 45 min (pACL), and endogenous peroxidase was blunted by incubating samples with 3% H2O2 for 10 min. After incubation with the primary antibody, slides were rinsed in PBS and signals were developed using the Vectastain Elite Kit and 3,3'-diaminobenzidine as a substrate for the peroxidase chromogenic reaction (Vector Laboratories, Burlingame, CA). At least 3 animals/group were evaluated.
Quantification of AcH4 positivity in acinar cells in Kras WT and KPCY mice (n=5/ group) was performed in a semi-automated manner using ImageJ software. For each animal, 5 to 10 images from different, non-overlapping microscopic fields were analyzed. Images containing malignant or pre-malignant lesions were excluded from the analysis. Count of nuclei within each image was calculated using ImageJ while positive nuclei were scored by a blinded evaluator. Ductal and islet cells were excluded by the evaluator.
Immunohistochemistry and automated scoring for human gliomas
Cases were obtained from the University of Pennsylvania following approval from the institutional review board. All cases were de-identified prior to analysis and were contained in a previously well-characterized tissue microarray sections (Venneti et al., 2013a). The cohort consisted of a total of 56 glioma samples (4 WHO grade II oligodendrogliomas, 5 grade III anaplastic oligodendrogliomas, 6 grade II oligoastrocytomas, 4 grade I pilocytic astrocytomas, 4 grade II diffuse astrocytomas, 9 grade III anaplastic astrocytomas, 14 grade IV glioblastomas, 2 grade I gangliogliomas and 8 grade II ependymomas). A neuropathologist evaluated all cases. Immunohistochemical studies and quantification were performed as previously described (Venneti et al., 2013b). Details may be found in Supplement.
Immunohistochemistry and scoring of human prostate tumors
Cases were obtained from the University of Pennsylvania following institutional review board approval. TMAs were assembled from primary tumors from 49 patients with either metastatic (n=25) or localized (n=24) disease. 5 μm sections from formalin-fixed paraffin-embedded (FFPE) TMA (Tissue Microarray) tissue blocks were prepared for immunohistochemical stain. Details of staining and scoring provided in Supplement.
Statistical Analyses
Student's 2-tailed t-tests were used for all analyses directly comparing 2 datasets. For in vitro assay testing AcCoA and CoASH effects on histone acetylation, repeated measures one-way ANOVA was used, with post test for linear trend. For correlations shown for human IHC, Pearson's product-moment correlation coefficient (Pearson's r) and corresponding 2-tailed significance (p value) was determined.
Supplementary Material
HIGHLIGHTS.
Acetyl-CoA: CoA ratio is glucose-sensitive and regulates histone acetylation levels
Oncogenic Akt expression promotes elevated histone acetylation in vitro and in vivo
ACLY phosphorylation by Akt enables sustained histone acetylation in low glucose
Histone acetylation and pAkt(Ser473) levels correlate significantly in human tumors
ACKNOWLEDGEMENTS
This work was supported by grants from the Pew Charitable Trusts (Biomedical Scholar Award), the Elsa U. Pardee Foundation, the American Diabetes Association (7-12-JF-59) and University of Pennsylvania Start-up Funds, to KEW. IAB was supported by NIH grants P30ES0138328 and T32ES019851. BAG acknowledges funding from the National Science Foundation Early Faculty CAREER award and an NIH Innovator grant (DP2OD007447) from the Office of the Director. KJW acknowledges R21-DK098769 and the University of Pennsylvania Diabetes Research Center (DRC) P30-DK19525. We thank George Belka for assistance with mammary tissue sections, as well as Dan Martinez and Li-Ping Wang for technical assistance with human tissue IHC. We thank the Functional Genomics Core at DRC (P30-DK19525) for RNA-sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS JVL performed and analyzed experiments in glioblastoma cells. AC performed and analyzed experiments using mouse tissues and cells from pancreas. SS performed and analyzed experiments in prostate cancer cells and assisted in other analyses. EJ provided technical assistance. NWS and AJW performed LC-MS metabolite analyses and NSW, AJW, and IAB analyzed data. SW and MDF analyzed human prostate cancer data. SV and AJ generated human glioma TMA and SV analyzed data. SL performed histone MS and ZFY and BAG analyzed data. NMA and BZS provided mouse pancreas sections and assisted in data interpretation. LAC provided mouse mammary tissue sections and assisted in data interpretation. HWL and KJW performed RNA-seq data analysis and clustering. NBH and TRR provided prostate cancer patient outcome data. KEW conceived and guided study, analyzed data, and wrote manuscript. All authors reviewed data and provided feedback on the manuscript.
REFERENCES
- Albaugh BN, Arnold KM, Denu JM. KAT(ching) metabolism by the tail: insight into the links between lysine acetyltransferases and metabolism. Chembiochem. 2011;12:290–298. doi: 10.1002/cbic.201000438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad N, Zahnow CA, Rudin CM, Baylin SB. The future of epigenetic therapy in solid tumours--lessons from the past. Nat Rev Clin Oncol. 2013;10:256–266. doi: 10.1038/nrclinonc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu SS, Blair IA. SILEC: a protocol for generating and using isotopically labeled coenzyme A mass spectrometry standards. Nat Protoc. 2012;7:1–12. doi: 10.1038/nprot.2011.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu SS, Mesaros C, Gelhaus SL, Blair IA. Stable isotope labeling by essential nutrients in cell culture for preparation of labeled coenzyme A and its thioesters. Anal Chem. 2011;83:1363–1369. doi: 10.1021/ac1027353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- Bianco-Miotto T, Chiam K, Buchanan G, Jindal S, Day TK, Thomas M, Pickering MA, O'Loughlin MA, Ryan NK, Raymond WA, Horvath LG, Kench JG, Stricker PD, Marshall VR, Sutherland RL, Henshall SM, Gerald WL, Scher HI, Risbridger GP, Clements JA, Butler LM, Tilley WD, Horsfall DJ, Ricciardelli C. Global levels of specific histone modifications and an epigenetic gene signature predict prostate cancer progression and development. Cancer Epidemiol Biomarkers Prev. 2010;19:2611–2622. doi: 10.1158/1055-9965.EPI-10-0555. [DOI] [PubMed] [Google Scholar]
- Cai L, Sutter BM, Li B, Tu BP. Acetyl-CoA induces cell growth and proliferation by promoting the acetylation of histones at growth genes. Mol Cell. 2011;42:426–437. doi: 10.1016/j.molcel.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervona Y, Costa M. Histone modifications and cancer: biomarkers of prognosis? Am J Cancer Res. 2012;2:589–597. [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Donohoe DR, Collins LB, Wali A, Bigler R, Sun W, Bultman SJ. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol Cell. 2012;48:612–626. doi: 10.1016/j.molcel.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg T, Schroeder S, Andryushkova A, Pendl T, Kuttner V, Bhukel A, Marino G, Pietrocola F, Harger A, Zimmermann A, Moustafa T, Sprenger A, Jany E, Buttner S, Carmona-Gutierrez D, Ruckenstuhl C, Ring J, Reichelt W, Schimmel K, Leeb T, Moser C, Schatz S, Kamolz LP, Magnes C, Sinner F, Sedej S, Frohlich KU, Juhasz G, Pieber TR, Dengjel J, Sigrist SJ, Kroemer G, Madeo F. Nucleocytosolic depletion of the energy metabolite acetyl-coenzyme a stimulates autophagy and prolongs lifespan. Cell Metab. 2014;19:431–444. doi: 10.1016/j.cmet.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB. Akt stimulates aerobic glycolysis in cancer cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- Evertts AG, Zee BM, Dimaggio PA, Gonzales-Cope M, Coller HA, Garcia BA. Quantitative dynamics of the link between cellular metabolism and histone acetylation. J Biol Chem. 2013;288:12142–12151. doi: 10.1074/jbc.M112.428318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fendt SM, Bell EL, Keibler MA, Olenchock BA, Mayers JR, Wasylenko TM, Vokes NI, Guarente L, Vander Heiden MG, Stephanopoulos G. Reductive glutamine metabolism is a function of the alpha-ketoglutarate to citrate ratio in cells. Nat Commun. 2013;4:2236. doi: 10.1038/ncomms3236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res. 2009;37:3969–3980. doi: 10.1093/nar/gkp270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gameiro PA, Yang J, Metelo AM, Perez-Carro R, Baker R, Wang Z, Arreola A, Rathmell WK, Olumi A, Lopez-Larrubia P, Stephanopoulos G, Iliopoulos O. In vivo HIF-mediated reductive carboxylation is regulated by citrate levels and sensitizes VHL-deficient cells to glutamine deprivation. Cell Metab. 2013;17:372–385. doi: 10.1016/j.cmet.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 2011;36:108–116. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther EJ, Belka GK, Wertheim GB, Wang J, Hartman JL, Boxer RB, Chodosh LA. A novel doxycycline-inducible system for the transgenic analysis of mammary gland biology. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2002;16:283–292. doi: 10.1096/fj.01-0551com. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hatzivassiliou G, Zhao F, Bauer DE, Andreadis C, Shaw AN, Dhanak D, Hingorani SR, Tuveson DA, Thompson CB. ATP citrate lyase inhibition can suppress tumor cell growth. Cancer Cell. 2005;8:311–321. doi: 10.1016/j.ccr.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Kaelin WG, Jr., McKnight SL. Influence of metabolism on epigenetics and disease. Cell. 2013;153:56–69. doi: 10.1016/j.cell.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katada S, Imhof A, Sassone-Corsi P. Connecting threads: epigenetics and metabolism. Cell. 2012;148:24–28. doi: 10.1016/j.cell.2012.01.001. [DOI] [PubMed] [Google Scholar]
- Killian JK, Kim SY, Miettinen M, Smith C, Merino M, Tsokos M, Quezado M, Smith WI, Jr., Jahromi MS, Xekouki P, Szarek E, Walker RL, Lasota J, Raffeld M, Klotzle B, Wang Z, Jones L, Zhu Y, Wang Y, Waterfall JJ, O'Sullivan MJ, Bibikova M, Pacak K, Stratakis C, Janeway KA, Schiffman JD, Fan JB, Helman L, Meltzer PS. Succinate dehydrogenase mutation underlies global epigenomic divergence in gastrointestinal stromal tumor. Cancer Discov. 2013;3:648–657. doi: 10.1158/2159-8290.CD-13-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurdistani SK. Histone modifications as markers of cancer prognosis: a cellular view. Br J Cancer. 2007;97:1–5. doi: 10.1038/sj.bjc.6603844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer MR, Fry CJ, Peterson CL, Denu JM. Modulating acetyl-CoA binding in the GCN5 family of histone acetyltransferases. J Biol Chem. 2002;277:27337–27344. doi: 10.1074/jbc.M203251200. [DOI] [PubMed] [Google Scholar]
- Letouze E, Martinelli C, Loriot C, Burnichon N, Abermil N, Ottolenghi C, Janin M, Menara M, Nguyen AT, Benit P, Buffet A, Marcaillou C, Bertherat J, Amar L, Rustin P, De Reynies A, Gimenez-Roqueplo AP, Favier J. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell. 2013;23:739–752. doi: 10.1016/j.ccr.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Lino MM, Merlo A. PI3Kinase signaling in glioblastoma. J Neurooncol. 2011;103:417–427. doi: 10.1007/s11060-010-0442-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono Gentile T, Lu C, Lodato PM, Tse S, Olejniczak SH, Witze ES, Thompson CB, Wellen KE. DNMT1 Is Regulated by ATP-Citrate Lyase and Maintains Methylation Patterns during Adipocyte Differentiation. Mol Cell Biol. 2013;33:3864–3878. doi: 10.1128/MCB.01495-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Losman JA, Kaelin WG., Jr. What a difference a hydroxyl makes: mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Thompson CB. Metabolic regulation of epigenetics. Cell Metab. 2012;16:9–17. doi: 10.1016/j.cmet.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, Wellen KE, O'Rourke DM, Berger SL, Chan TA, Levine RL, Mellinghoff IK, Thompson CB. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum JJ, Bauer DE, Kong M, Harris MH, Li C, Lindsten T, Thompson CB. Growth factor regulation of autophagy and cell survival in the absence of apoptosis. Cell. 2005;120:237–248. doi: 10.1016/j.cell.2004.11.046. [DOI] [PubMed] [Google Scholar]
- Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. 2012;23:560–566. doi: 10.1016/j.tem.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino G, Pietrocola F, Eisenberg T, Kong Y, Malik SA, Andryushkova A, Schroeder S, Pendl T, Harger A, Niso-Santano M, Zamzami N, Scoazec M, Durand S, Enot DP, Fernandez AF, Martins I, Kepp O, Senovilla L, Bauvy C, Morselli E, Vacchelli E, Bennetzen M, Magnes C, Sinner F, Pieber T, Lopez-Otin C, Maiuri MC, Codogno P, Andersen JS, Hill JA, Madeo F, Kroemer G. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol Cell. 2014;53:710–725. doi: 10.1016/j.molcel.2014.01.016. [DOI] [PubMed] [Google Scholar]
- Martinez-Pastor B, Cosentino C, Mostoslavsky R. A tale of metabolites: the cross-talk between chromatin and energy metabolism. Cancer Discov. 2013;3:497–501. doi: 10.1158/2159-8290.CD-13-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBrian MA, Behbahan IS, Ferrari R, Su T, Huang TW, Li K, Hong CS, Christofk HR, Vogelauer M, Seligson DB, Kurdistani SK. Histone acetylation regulates intracellular pH. Mol Cell. 2013;49:310–321. doi: 10.1016/j.molcel.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migita T, Narita T, Nomura K, Miyagi E, Inazuka F, Matsuura M, Ushijima M, Mashima T, Seimiya H, Satoh Y, Okumura S, Nakagawa K, Ishikawa Y. ATP citrate lyase: activation and therapeutic implications in non-small cell lung cancer. Cancer Res. 2008;68:8547–8554. doi: 10.1158/0008-5472.CAN-08-1235. [DOI] [PubMed] [Google Scholar]
- Morris J.P.t., Wang SC, Hebrok M. KRAS, Hedgehog, Wnt and the twisted developmental biology of pancreatic ductal adenocarcinoma. Nat Rev Cancer. 2010;10:683–695. doi: 10.1038/nrc2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapova IA, El-Maghrabi MR, Doronin SV, Benjamin WB. Phosphorylation of recombinant human ATP:citrate lyase by cAMP-dependent protein kinase abolishes homotropic allosteric regulation of the enzyme by citrate and increases the enzyme activity. Allosteric activation of ATP:citrate lyase by phosphorylated sugars. Biochemistry. 2000;39:1169–1179. doi: 10.1021/bi992159y. [DOI] [PubMed] [Google Scholar]
- Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, Reichert M, Beatty GL, Rustgi AK, Vonderheide RH, Leach SD, Stanger BZ. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148:349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–1266. doi: 10.1038/nature03672. [DOI] [PubMed] [Google Scholar]
- Shen H, Laird PW. Interplay between the Cancer Genome and Epigenome. Cell. 2013;153:38–55. doi: 10.1016/j.cell.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Tu BP. Acetyl-CoA induces transcription of the key G1 cyclin CLN3 to promote entry into the cell division cycle in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2013;110:7318–7323. doi: 10.1073/pnas.1302490110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son J, Lyssiotis CA, Ying H, Wang X, Hua S, Ligorio M, Perera RM, Ferrone CR, Mullarky E, Shyh-Chang N, Kang Y, Fleming JB, Bardeesy N, Asara JM, Haigis MC, DePinho RA, Cantley LC, Kimmelman AC. Glutamine supports pancreatic cancer growth through a KRAS-regulated metabolic pathway. Nature. 2013;496:101–105. doi: 10.1038/nature12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, McCaffery JM, Irizarry RA, Boeke JD. Nucleocytosolic acetyl-coenzyme a synthetase is required for histone acetylation and global transcription. Mol Cell. 2006;23:207–217. doi: 10.1016/j.molcel.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Ulanovskaya OA, Zuhl AM, Cravatt BF. NNMT promotes epigenetic remodeling in cancer by creating a metabolic methylation sink. Nat Chem Biol. 2013;9:300–306. doi: 10.1038/nchembio.1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG. Targeting cancer metabolism: a therapeutic window opens. Nat Rev Drug Discov. 2011;10:671–684. doi: 10.1038/nrd3504. [DOI] [PubMed] [Google Scholar]
- Venneti S, Felicella MM, Coyne T, Phillips JJ, Gorovets D, Huse JT, Kofler J, Lu C, Tihan T, Sullivan LM, Santi M, Judkins AR, Perry A, Thompson CB. Histone 3 lysine 9 trimethylation is differentially associated with isocitrate dehydrogenase mutations in oligodendrogliomas and high-grade astrocytomas. J Neuropathol Exp Neurol. 2013a;72:298–306. doi: 10.1097/NEN.0b013e3182898113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venneti S, Garimella MT, Sullivan LM, Martinez D, Huse JT, Heguy A, Santi M, Thompson CB, Judkins AR. Evaluation of Histone 3 Lysine 27 Trimethylation (H3K27me3) and Enhancer of Zest 2 (EZH2) in Pediatric Glial and Glioneuronal Tumors Shows Decreased H3K27me3 in H3F3A K27M Mutant Glioblastomas. Brain Pathol. 2013b;23:558–564. doi: 10.1111/bpa.12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Lu C, Mancuso A, Lemons JM, Ryczko M, Dennis JW, Rabinowitz JD, Coller HA, Thompson CB. The hexosamine biosynthetic pathway couples growth factor-induced glutamine uptake to glucose metabolism. Genes Dev. 2010;24:2784–2799. doi: 10.1101/gad.1985910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, Zhao S, Ye D, Xiong Y, Guan KL. Inhibition of alpha-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev. 2012;26:1326–1338. doi: 10.1101/gad.191056.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H, Kimmelman AC, Lyssiotis CA, Hua S, Chu GC, Fletcher-Sananikone E, Locasale JW, Son J, Zhang H, Coloff JL, Yan H, Wang W, Chen S, Viale A, Zheng H, Paik JH, Lim C, Guimaraes AR, Martin ES, Chang J, Hezel AF, Perry SR, Hu J, Gan B, Xiao Y, Asara JM, Weissleder R, Wang YA, Chin L, Cantley LC, DePinho RA. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670. doi: 10.1016/j.cell.2012.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Johnson JL, Hanigan CL, Locasale JW. Interactions between epigenetics and metabolism in cancers. Front Oncol. 2012;2:163. doi: 10.3389/fonc.2012.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.