Significance
Benzenediol lactone (BDL) polyketides are privileged structures whose various members bind to distinct receptors or modulate the heat shock response and the immune system. BDLs are biosynthesized by collaborating polyketide synthase enzyme pairs in fungi. Coexpressing random heterocombinations of these enzymes from different BDL biosynthetic pathways in yeast cells is shown here to lead to the one-pot, one-step combinatorial biosynthesis of structurally diverse polyketides in practical amounts. Combinatorial biosynthesis promises to generate a novel unexplored source of bioactive molecules in an environmentally sustainable, economical, and inherently scalable manner. Broadening the medicinally relevant chemical space of polyketides by such methods will provide unnatural products as valuable entry points for drug discovery and development.
Keywords: secondary metabolites, fungal genetics
Abstract
Combinatorial biosynthesis aspires to exploit the promiscuity of microbial anabolic pathways to engineer the synthesis of new chemical entities. Fungal benzenediol lactone (BDL) polyketides are important pharmacophores with wide-ranging bioactivities, including heat shock response and immune system modulatory effects. Their biosynthesis on a pair of sequentially acting iterative polyketide synthases (iPKSs) offers a test case for the modularization of secondary metabolic pathways into “build–couple–pair” combinatorial synthetic schemes. Expression of random pairs of iPKS subunits from four BDL model systems in a yeast heterologous host created a diverse library of BDL congeners, including a polyketide with an unnatural skeleton and heat shock response-inducing activity. Pairwise heterocombinations of the iPKS subunits also helped to illuminate the innate, idiosyncratic programming of these enzymes. Even in combinatorial contexts, these biosynthetic programs remained largely unchanged, so that the iPKSs built their cognate biosynthons, coupled these building blocks into chimeric polyketide intermediates, and catalyzed intramolecular pairing to release macrocycles or α-pyrones. However, some heterocombinations also provoked stuttering, i.e., the relaxation of iPKSs chain length control to assemble larger homologous products. The success of such a plug and play approach to biosynthesize novel chemical diversity bodes well for bioprospecting unnatural polyketides for drug discovery.
Encompassing one of the largest classes of structurally diverse small molecule natural products, polyketides have provided multiple clinically useful drug classes that save lives (1–3). Natural polyketides from diverse microorganisms also deliver novel scaffolds that can be exploited in drug discovery programs by semisynthetic modification and combined chemical and biosynthetic approaches (4, 5) and as inspiration for total synthesis and combinatorial chemistry (6, 7). A complementary approach is combinatorial biosynthesis, which strives to reengineer biosynthetic pathways to generate novel polyketide scaffolds by one-pot, one-step synthesis via fermentation of recombinant microorganisms.
Among fungal polyketides, benzenediol lactones (BDLs) offer rich pharmacophores with an extraordinary range of biological activities (8). They are defined by a 1,3-benzenediol moiety bridged by a macrocyclic lactone ring (9). Among BDLs, resorcylic acid lactones (RALs) display a C2–C7 connectivity, whereas dihydroxyphenylacetic acid lactones (DALs) feature a C3–C8 bond. Monocillin II (1; Fig. 1) and their congeners (radicicol and the pochonins) are RALs with 14-membered rings (RAL14) that are specific inhibitors of heat shock protein 90 (Hsp90) (9, 10). Inhibition of Hsp90 promotes the degradation of oncogenic client proteins and leads to the combinatorial blockade of multiple cancer-causing pathways. Resorcylides and lasiodiplodins are RALs with 12-membered macrocycles (RAL12): these phytotoxins display mineralocorticoid receptor antagonist and prostaglandin biosynthesis inhibitory activities in animals. 10,11-dehydrocurvularin (7; Fig. 1) is a DAL with a 12-membered ring (DAL12) that modulates the heat shock response and the immune system (8, 9).
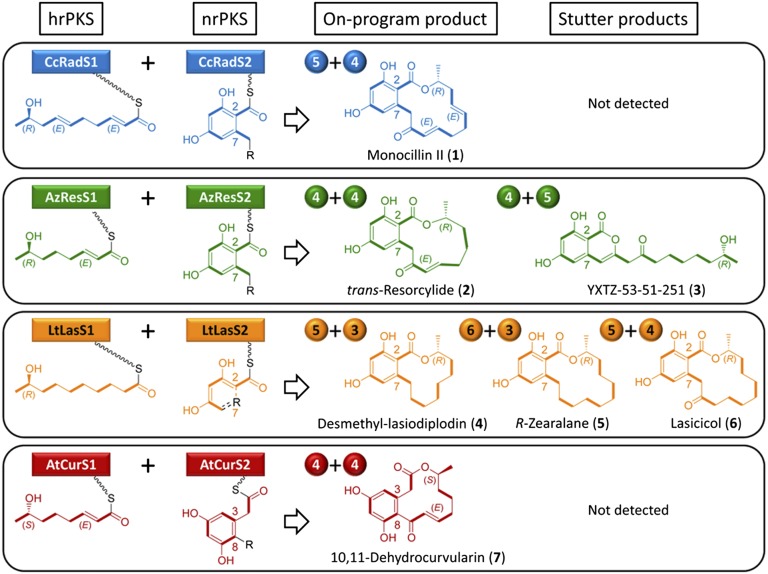
Fig. 1.
Biosynthesis of natural benzenediol lactones. Biosynthetic assembly of monocillin II, trans-resorcylide, desmethyl-lasiodiplodin, and 10,11-dehydrocurvularin (the “on-program” main metabolites) in recombinant S. cerevisiae BJ5464-NpgA (24, 40) strains by native BDLSs (12, 14, 25). R indicates the hrPKS-generated priming unit that is elaborated by the nrPKS. Numerals in the colored spheres indicate the number of malonate-derived C2 units (C−C bonds shown in bold) incorporated into the polyketide chain by the hrPKS vs. the nrPKS (“division of labor” or “split” by the BDLS: e.g., 5+4 indicates a pentaketide extended by four more malonate units). Stutter products are minor metabolites produced by extra extension cycles resulting in irregular “splits” (25).
BDL scaffolds are biosynthesized by pairs of collaborating, sequentially acting iterative polyketide synthases (iPKSs) (3) forming quasi-modular BDL synthases (BDLSs) (Fig. 1) (11–14). Each of the BDLS subunits catalyze recursive, decarboxylative Claisen condensations of malonyl-CoA using a single core set of ketoacyl synthase (KS), acyl transferase (AT), and acyl carrier protein (ACP) domains. BDL assembly initiates on a highly reducing iPKS (hrPKS) that produces a short chain carboxylic acid priming unit for a second, nonreducing iPKS (nrPKS). The length of the priming unit is set by the KS domain of the hrPKS, whereas the distinctive redox pattern and the configuration at each stereocenter is determined by the ketoreductase (KR), dehydratase (DH), and enoyl reductase (ER) domains that reduce the nascent β-keto groups after each condensation step according to a cryptic biosynthetic program (3). A direct handover of the priming unit from the hrPKS is catalyzed by the starter unit:ACP transacylase (SAT) domain of the nrPKS (15). After a set number of further elongation cycles without reduction, the highly reactive polyketide intermediate is guided by the product template (PT) domain of the nrPKS toward a programmed, regiospecific first ring closure (16). This aldol condensation yields a resorcylic acid moiety in the C2–C7 register or a dihydroxyphenylacetic acid group in the C8–C3 register (16–18). The last step of BDL scaffold biosynthesis is the release of the product, catalyzed by an O–C bond-forming thioesterase (TE) domain of the nrPKS. TE domains form the BDL macrolactone using the ω-1 alcohol as a nucleophile, but may use alternative nucleophiles such as the C9 enol to yield α-pyrones or water or alcohols from the media to form acyl resorcylic acids (ARAs), acyl dihydroxyphenylacetic acids (ADAs), and their esters (18–22). iPKSs that produce BDLs in the RAL14, RAL12, and DAL12 subclasses have been characterized and reconstituted both in vivo by heterologous expression in yeast and in vitro using isolated recombinant iPKS enzymes (11–14, 23–25). Domain exchanges among different BDLSs were used to decipher some of the programming rules of these enzymes and yielded a limited number of structurally diverse unnatural products (3, 18, 20, 21, 26, 27).
Combinatorial biosynthesis of polyketides is still in its infancy. Mix and match combinations of small, discrete type II PKSs from bacteria have yielded some novel scaffolds, but the outcome of the reactions proved difficult to predict and conceptualize. Engineering targeted changes in modular type I PKSs of prokaryotes generated small, focused libraries of conservative variants of selected bioactive scaffolds. However, these pioneering works with prokaryotic PKSs also yielded many unproductive combinations, suffered from greatly reduced product yields, and realized only a small fraction of the potential chemical space (28–31).
Large-scale combinatorial biosynthesis with fungal iPKSs has not yet been described. Nevertheless, fungal BDLs provide a unique opportunity for diversity-oriented combinatorial biosynthesis. This is because BDLSs from different organisms are orthologous enzymes that are nonetheless programmed to generate nonequivalent, structurally complex macrocyclic products using malonyl-CoA as their sole, shared precursor. In this work, we considered whether BDL biosynthesis may be refactored to a modular, parallel synthetic scheme in which individually programmed biosynthons are freely coupled and released after intramolecular cyclization, in analogy to a build–couple–pair strategy in combinatorial chemistry (7, 32). Further, we explored whether BDLS subunit heterocombinations also reveal differences in iPKS promiscuity and plasticity.
Results and Discussion
Plug and Play BDL Biosynthesis.
We have used four iPKS pairs as model systems: CcRadS1–CcRadS2 for the RAL14 monocillin II (a precursor of radicicol); AzResS1–AzResS2 and LtLasS1–LtLasS2 for the RAL12 resorcylides and lasiodiplodins, respectively; and AtCurS1–AtCurS2 for the DAL12 curvularins (Fig. 1) (12, 14, 25). Although originating from fungi of different Pezizomycotina classes, the hrPKSs on one hand and the nrPKSs on the other are orthologous and display identical domain organizations, raising hopes that the channeling of biosynthetic intermediates across systems may be feasible. Nevertheless, the BDLS subunits build characteristically different, orthogonal biosynthons (Fig. 1). Thus, CcRadS1 and LtLasS1 yield pentaketides with distinct redox patterns, whereas AzResS1 and AtCursS1 elaborate enantiomeric tetraketides differing only in the configuration of the ω-1 stereocenter. CcRadS2 and AzResS2 each extend their priming units with four additional malonyl-CoA units and catalyze aldol condensation in the C2–C7 register to yield a resorcylate carboxylic acid moiety (17, 18). Although LtLasS2 also assembles a resorcylate building block, it catalyzes only three extension cycles. Finally, AtCurS2 catalyzes four extension cycles but generates a dihydroxyphenylacetic acid moiety by a C8–C3 aldol reaction (14, 18). We have previously demonstrated the in vivo production of the native polyketide products of the cognate BDLS subunit pairs using compatible yeast expression vectors (14, 18, 25). In addition to the main on-program products, both the AzResS and the LtLasS systems yield stutter products when expressed in yeast (Fig. 1) (25). Stuttering involves the incorporation of an extra malonate unit by the hrPKS or the nrPKS, leading to an irregular division of labor or “split” between the BDLS subunits (25, 33). For the current study, we coexpressed the four hrPKSs and the four nrPKSs in every possible pairwise combination. If a subunit heterocombination failed to generate a polyketide (or only did so in minute quantities), we attempted to force the coupling by replacing the SAT domain of the nrPKS (15, 20, 34) with the one that is cognate to the hrPKS. Finally, the intramolecular pair step of the combinatorial synthetic scheme was provided by the nrPKS TE domains to yield a macrocycle or an α-pyrone. Alternatively, TEs may form ARA, ADA, or their esters by using water or alcohols from the media as the nucleophile (20).
RAL Formation with nrPKSs Catalyzing Four Extension Steps.
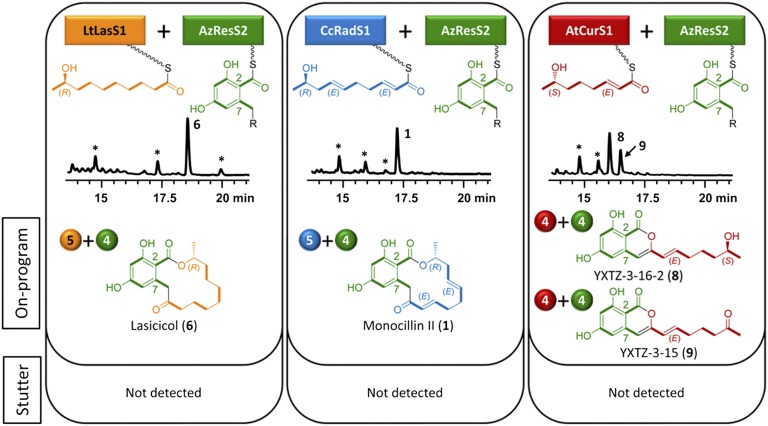
Pairing the nrPKSs AzResS2 or CcRadS2 with the LtLasS1 hrPKS provided the expected on-program RAL14 product, lasicicol (6), in good yields (∼8 mg/L with AzResS2 and 9 mg/L with CcRadS2; Figs. 2 and 3). Lasicicol was previously isolated as a minor stutter product of the heterologously expressed LtLasS system (25). Pairing AzResS2 with CcRadS1 afforded the on-program RAL14 product monocillin II (1) in a good yield (6 mg/L; Fig. 2). Efficient production of 1 and 6 indicates that AzResS2 is competent to transfer and extend not only tetraketide, but also pentaketide priming units from heterologous hrPKSs, and is capable of releasing the longer product by macrocycle formation. Variant priming unit redox patterns (presence or absence of double bonds at C2,C3 or at C6,C7) are not impediments for this enzyme either. Coupling AzResS2 with AtCurS1 provided the known isocoumarins YXTZ-3-16-2 (8, 5 mg/L) and YXTZ-3-15 (9, 3 mg/L; Fig. 2). Previously observed in low yields, 9 derives by a chance oxidation of the C15 alcohol of 8 by the yeast host (18). Although AzResS2 is competent to accept and extend either of the enantiomeric tetraketides produced by AzResS1 and AtCursS1, it is unable to form a macrocycle with the C7 alcohol in the (S) configuration. No stutter products were formed in AzResS2 heterocombinations, in contrast to fermentations with the cognate AzResS pair (Fig. 1 vs. Fig. 2).
Fig. 2.
Combinatorial biosynthesis using AzResS2. Biosynthesis of on-program and stutter products by S. cerevisiae BJ5464-NpgA (24, 40) coexpressing AzResS2 with heterologous BDL hrPKSs (12, 14, 25). Product structures are colored to indicate the origin of the assembled biosynthons. Stars over peaks in product profiles (HPLC traces recorded at 300 nm) indicate host-derived metabolites. See Fig. 1 for additional explanations.
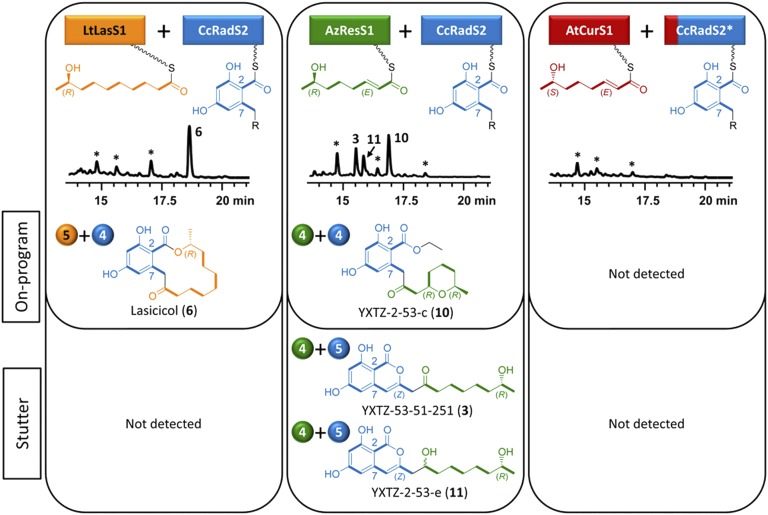
Fig. 3.
Combinatorial biosynthesis using CcRadS2. Biosynthesis of on-program and stutter products by S. cerevisiae BJ5464-NpgA (24, 40) coexpressing CcRadS2 with heterologous BDL hrPKSs (12, 14, 25). CcRadS2* and the split-color box indicates that SATCcRadS2 was replaced by SATAtCurS2 to facilitate coupling with AtCurS1. See Fig. 2 for additional explanations.
CcRadS2 provided an even more stark contrast for the utilization of the enantiomeric priming units of AzResS1 and AtCurS1 (Fig. 3). The C7(R) isomer was accepted from AzResS1 and extended efficiently to yield the on-program ARA ethyl ester YXTZ-2-53-c (10, 5 mg/L), even if the shorter product was apparently not compatible with macrocycle formation by TECcRadS2. To our surprise, stutter isocoumarin products with an apparent 4+5 split were also formed in good yields (3, 4 mg/L and 11, 3 mg/L), with the racemic 11 presumably derived from 3 by a chance reduction. Formation of small amounts of 3 was also detected with the cognate AzResS pair upon heterologous expression (25), but stuttering has not previously been observed with CcRadS2. Meanwhile, the isomeric (7S) tetraketide was not accepted by CcRadS2 from AtCurS1, even after replacement of the SATCcRadS2 domain with SATAtCurS2 (Fig. 3). Replacing the TE or the ACP–TE didomain of CcRadS2 with those of AtCurS2 in addition to the SAT exchange did not yield any product either (20). Only the substitution of the SAT–KS–AT tridomain of CcRadS2 for that of AtCurS2 allowed pairing with AtCurS1 to produce the ARA ethyl ester YXTZ-3-49-1 (20).
RAL Formation with an nrPKS Catalyzing Three Extension Cycles.
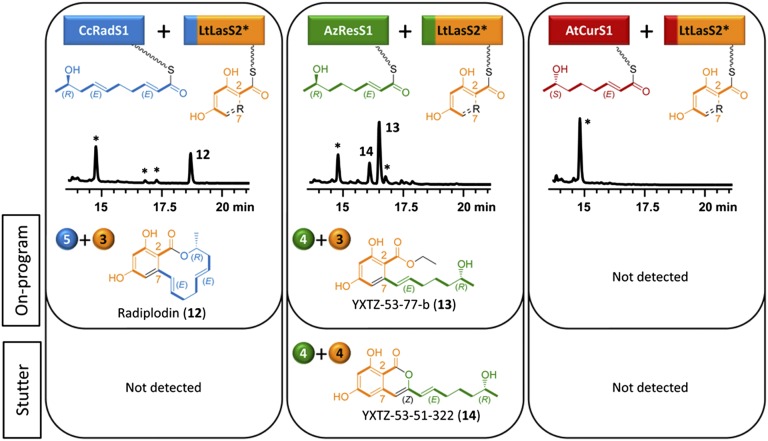
The SAT domain of LtLasS2 turned out to be a fastidious catalyst because no products formed on pairing this nrPKS with noncognate hrPKSs. However, the rest of the LtLasS2 chassis is more promiscuous, because SAT domain exchanges allowed product formation in two of the three subunit heterocombinations (Fig. 4). Thus, the expected on-program RAL12 product radiplodin (12) was recovered in moderate yields (3 mg/L) on pairing LtLasS2 with CcRadS1, indicating that the two extra double bonds in the priming unit (compared with the LtLasS1-generated starter) are not impediments for the extension or the macrocyclization steps. Stutter products were not detected with the CcRadS1–LtLasS2 pair. Challenging LtLasS2 with the enantiomeric tetraketides produced by AzResS1 and AtCurS1 proved illuminating, for it neatly mirrored the results obtained with CcRadS2 earlier (Fig. 4 vs. Fig. 3). Just like CcRadS2, LtLasS2 also produced large amounts of an on-program ARA ester (13, 8 mg/L) and a stutter α-pyrone (14, 3 mg/L, 4+4 split) when provided with the 7R tetraketide by AzResS1 (Fig. 4). Thus, a priming unit that is shorter than the native ones for CcRadS2 and LtLasS2 may still be suitable for chain extension but not for macrocycle formation. However, when the ω-1 stereocenter of this short priming unit is in the opposite configuration (as with the 7S tetraketide from AtCurS1), product formation was completely abolished, despite the replacement of SATLtLasS2 with SATAtCurS2 (Fig. 4). Collectively, these experiments stress the importance of the ω-1 stereocenter beyond determining the configuration of the exocyclic methyl group in BDLs. Confronted with a priming unit with the alcohol in the wrong configuration, the nrPKS chassis may release an α-pyrone instead of a macrocycle (AzResS2; Fig. 2), or abolish polyketide chain extension and/or product release (CcRadS2 and LtLasS2; Figs. 3 and 4).
Fig. 4.
Combinatorial biosynthesis using LtLasS2. Biosynthesis of on-program and stutter products by S. cerevisiae BJ5464-NpgA (24, 40) coexpressing LtLasS2 with heterologous BDL hrPKSs (12, 14, 25). LtLasS2* in a split-color box indicates replacement of SATLtLasS2 with the SAT cognate for the heterologous hrPKS (e.g., SATLtLasS2 replaced by SATCcRadS2 to facilitate collaboration of LtLasS2 with CcRadS1). See Fig. 2 for additional explanations.
DAL Formation with an nrPKS Catalyzing Four Extension Cycles.
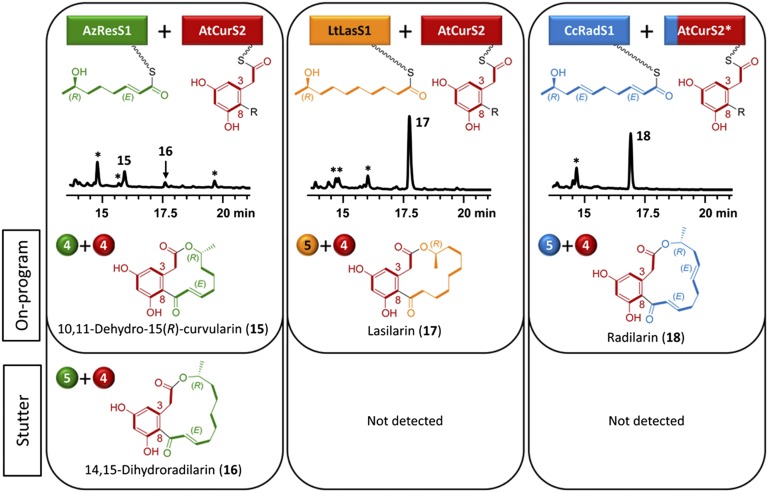
The configuration of the ω-1 alcohol proved similarly important in pairing AtCurS2 with AzResS1 (Fig. 5). Here, the expected on-program DAL12 product, epi-dehydrocurvularin (15) formed in low yields (0.8 mg/L), accompanied by small amounts of the stutter (5+4 split) DAL14 product 14,15-dihydroradilarin (16, 0.1 mg/L). Thus, the unfavorable configuration of the stereocenter depressed the yield but did not obstruct macrocycle formation. Stuttering of AzResS1, apparent during the formation of 16, has not been detected with the cognate AzResS pair (25). Interestingly, neither the R configuration nor the increased length of the priming unit proved to be a barrier to the formation of the expected on-program DAL14 products with LtLasS1 (lasilarin 17, 10 mg/L) and with CcRadS1 (radilarin 18, 9 mg/L; Fig. 5). The coupling of AtCurS2 with CcRadS1 needed to be enforced by an SAT exchange, although trace amounts of 18 also formed without this manipulation (20). No stutter or nonmacrocyclic products were detected with either subunit heterocombinations.
Fig. 5.
Combinatorial biosynthesis using AtCurS2. Biosynthesis of on-program and stutter products by S. cerevisiae BJ5464-NpgA (24, 40) coexpressing AtCurS2 with heterologous BDL hrPKSs (12, 14, 25). AtCurS2* in a split-color box indicates replacement of SATAtCurS2 with SATCcRadS2. See Fig. 2 for additional explanations.
Biological Activities.
Novel BDLs and their congeners isolated in this study were evaluated for their in vitro inhibition of cell proliferation against a panel of five human cancer cell lines and for their heat shock-inducing activity against a reporter cell line (Table 1) (35, 36). Only radilarin (18) with a DAL14 skeleton showed significant cytotoxicity, although its potency remained below that of the comparable natural DAL12 product 10,11-dehydrocurvularin (7). In contrast, radilarin displayed a more potent heat shock response modulatory activity than dehydrocurvularin, although both compounds were significantly less active than monocillin I, used as the positive control (Table 1). Although the RAL14 monocillin I and its congeners (radicicol and the pochonins) inhibit Hsp90 (10, 37), the mechanism of heat shock modulation by the DAL12 7 has not been clarified (36). The two additional DAL14 compounds isolated here, 14,15-dihydroradilarin (16) and lasilarin (17), showed no cytotoxicity or heat shock induction activity at 5 μM, stressing the importance of the 14(E) double bond for both activities. Similarly, epi-dehydrocurvularin (15) showed no activities in either assays at 5 μM, indicating that the configuration of the exocyclic methyl is critical for inhibition of the targets.
Table 1.
Biological activities of selected compounds
| Compound | Cytotoxicity | HSIA | ||||
| PC3M | NCI-H460 | SF-268 | MCF-7 | MDA-MB-231 | ||
| MON* | NT | 0.4 ± 0.01 | 0.3 ± 0.01 | 0.3 ± 0.01 | NT | 732 ± 58 |
| DOX* | 0.3 ± 0.01 | 0.3 ± 0.01 | 0.6 ± 0.01 | 0.4 ± 0.01 | 0.5 ± 0.10 | NT |
| 7 | 1.7 ± 0.06 | 1.4 ± 0.09 | 1.4 ± 0.2 | 2.0 ± 0.12 | 2.9 ± 0.40 | 314 ± 14 |
| 18 | 3.0 ± 0.11 | 2.3 ± 0.04 | 3.1 ± 0.08 | 2.7 ± 0.14 | 2.7 ± 0.14 | 815 ± 69 |
Cytotoxic activities are shown as IC50 values ± SD in μM vs. the following human cell lines: PC3M, metastatic prostate adenocarcinoma; NCI-H460, non–small-cell lung cancer; SF-268, central nervous system glioma; MCF-7, breast cancer; MDA-MB-231, metastatic breast adenocarcinoma. Heat-shock induction activities (HSIA) are shown as the means ± SDs in percentages of the negative control (DMSO). Compounds were tested at 5.0 μM except the positive controls (MON, monocillin I and DOX, doxorubicin), evaluated at 0.5 μM (*). NT, not tested.
Composite Programming of BDLS Subunit Heterocombinations.
Random subunit heterocombinations revealed a surprising promiscuity and flexibility of the model BDLSs (Supporting Information). Regardless of the sizes of their native priming units, all four nrPKSs accepted and processed pentaketide biosynthons. In particular, the flexible 9(R)-hydroxydecanoic acid priming unit from LtLasS1 was universally accepted by all nrPKSs without necessitating SAT domain exchanges, and supported the formation of RAL14, RAL12, and DAL14 product skeletons in uniformly good yields (Supporting Information). Neither the length nor the altered redox pattern of the pentaketide priming units influenced the innate programming of the heterologous nrPKSs, not even those expecting a tetraketide primer, with the number of extension cycles, the regioselectivity of aldol condensations, or the release of products as macrocycles remaining on-program. Importantly, stutter products were not detected in any subunit heterocombinations with pentaketide-forming hrPKSs and were present in trace amounts only with the native LtLasS pair (Figs. 2–5).
In contrast, shorter-than-expected priming units posed an increased challenge. First, although the AzResS1-derived tetraketide was accepted by the pentaketide-expecting nrPKSs CcRadS2 and LtLasS2, this priming unit provoked these nrPKSs to stutter profusely (Figs. 3 and 4 and Supporting Information). We hypothesize that the assembly of stutter products by iPKSs is under kinetic control (38). In combinatorial contexts, the intrinsic rate of the ACP-to-KS retrotransfer of a shorter intermediate for an extra (stutter) round of ketide homologation can surpass the rate of the on-program processing of that unexpected substrate by a downstream enzyme (SAT for the priming unit on the hrPKS, and PT and TE for the assembled polyketide on the nrPKS). Next, nrPKSs expecting a pentaketide primer were unable to form ring-contracted macrocycles with a tetraketide starter, presumably because shorter-than-expected polyketides are unsuitable substrates for macrocycle formation by the TEs. Last, the AtCurS1-derived tetraketide with the wrong ω-1 stereocenter configuration proved especially problematic, with the pentaketide-expecting nrPKSs refusing this priming unit despite SAT domain exchanges, and AzResS2 generating only α-pyrone products (Figs. 2–4 and Supporting Information). Interestingly, the configuration of the ω-1 stereocenter is far less important for TEAtCurS2: this enzyme is able to use intermediates featuring either the native (S) or the unexpected (R) configuration for macrocycle formation, albeit at different yields (Fig. 5).
Conclusions.
Using BDL assembly as a model, we demonstrate for the first time, to our knowledge, a working example of a diversity-oriented combinatorial biosynthetic scheme for fungal iPKSs. We show that subunit heterocombinations of four orthologous BDLSs allow the efficient biosynthesis of structurally complex macrocyclic polyketides and their congeners, with 14 of the possible 16 combinations (including the native pairs) producing isolable amounts of polyketides in the heterologous host (Supporting Information). Because the four BDLS systems originated from distant fungal lineages and display orthogonal product specificities (RAL14, RAL12, and DAL12), their productive collaboration to assemble diverse molecules in good yields was by no means guaranteed. Given the success of these experiments, we expect that extending our combinatorial matrix with additional, native, or chimeric BDLSs embodying different biosynthetic programs will substantially widen the product spectrum while at the same time reveal the innate programming of iPKSs. Thus, successful transfer of heterologous priming units between noncognate enzyme pairs revealed that SAT domains are polyspecific (1, 33). Nevertheless, SAT domain swaps still proved necessary in some cases to enhance system efficiency by reinforcing BDLS subunit coupling (34). The rest of the nrPKS chassis turned out to be surprisingly flexible in elaborating foreign biosynthons while remaining mostly faithful to their intrinsic biosynthetic programs. However, shorter-than-expected biosynthons provoked stuttering, highlighting a possible kinetic competition between chain extension vs. product release (25, 39). The decision gating role of the TE domains (20) was also manifest in substrate-dependent shifts in product release modes, including the formation of macrocycles, α-pyrones, ARA, ADA, and their esters. The unexpected success of the modularization of BDL biosynthesis suggests that diversity-oriented combinatorial biosynthesis will be able to extend the natural product chemical space in a time- and resource-efficient manner. Our work provides a biosynthetic tool to generate unnatural polyketides as an unexplored source of chemical diversity and novelty, ready to be exploited for drug discovery.
Materials and Methods
Strains and Culture Conditions.
S. cerevisiae BJ5464-NpgA (MATα ura3-52 his3-Δ200 leu2-Δ1 trp1 pep4::HIS3 prb1 Δ1.6R can1 GAL) (40, 41) served as the host for expression vectors based on YEpADH2p-FLAG-URA and YEpADH2p-FLAG-TRP (14, 18, 20, 25). Construction of expression vectors are described in SI Materials and Methods. Recombinant yeast strains were maintained at 30 °C on synthetic complete agar medium [0.67% yeast nitrogen base, 2% (wt/vol) glucose, 1.5% (wt/vol) agar, and 0.72 g/L Trp/Ura DropOut supplement].
Production and Chemical Characterization of Polyketides.
Three to five independent S. cerevisiae transformants were analyzed for polyketide production, and fermentations with representative isolates were repeated at least three times. Extraction of polyketides, analysis of extracts by reversed phase HPLC, scaled-up cultivation of yeast strains, and isolation of products followed previous protocols (14, 18, 20, 25). Low-resolution mass measurements were done on an Agilent 6130 Single Quad LC-MS. Optical rotations were recorded on a Rudolph Autopol IV polarimeter. Circular dichroism (CD) spectra were acquired with a JASCO J-810 instrument. 1H, 13C, and 2D NMR (homonuclear correlation, heteronuclear single quantum coherence, and heteronuclear multiple-bond correlation) spectra were obtained in CD3OD or C5D5N on a JEOL ECX-300 spectrometer. See SI Materials and Methods for details.
Biological Assays.
Cytotoxicity was evaluated by measuring the increase of fluorescence on reduction of resazurin (AlamarBlue) using human non–small-cell lung cancer (NCI-H460), central nervous system glioma (SF-268), breast cancer (MCF-7), metastatic prostate adenocarcinoma (PC-3M), and human metastatic breast adenocarcinoma (MDA-MB-231) cell lines. Vehicle (DMSO) and positive controls (the chemotherapeutic drug doxorubicin at 0.5 μM), and serial dilutions of test compounds (starting at 5 μM) were evaluated in triplicate, and the assays were repeated twice. Mean fluorescence intensity compared with vehicle controls served as a measure of relative cell viability, as described previously (35). The semiquantitative heat-shock induction assay (HSIA) used mouse fibroblasts stably transduced with a reporter construct encoding the enhanced GFP under the transcriptional control of a minimal consensus heat shock element (36, 42). Test compounds at 5 μM were measured in triplicate, with DMSO vehicle as the negative control and monocillin I as the positive control (tested at 0.5 μM). Fluorescence was determined on an Analyst AD (LJL Biosystems) plate reader (excitation, 485 nm; emission, 525 nm). Relative fluorescence intensities were normalized to the DMSO control values and reported as percentages; HSIA activities >250% are considered significant.
Supplementary Material
Acknowledgments
We thank N. A. DaSilva (University of California, Irvine) for S. cerevisiae BJ5464-NpgA, Y. Tang (University of California, Los Angeles) for the YEpADH2p vectors, and M. X. Liu (University of Arizona, Tucson) for assistance with bioassays. This work was supported by National Science Foundation Grant MCB-0948751 (to I.M.), National Institutes of Health Grants AI065357 RM DP 008 (to J.Z.) and R01 CA090265 (to A.A.L.G.), American Heart Association Grant 09SDG2060080 (to J.Z.), the Chinese Academy of Agricultural Sciences Elite Youth Program (Y.X.), National Research and Development Project of Transgenic Crops of China Grant 2013ZX08012-001 (to W.Z.), and National Basic Research Program of China Grant 2013CB733903 (to M.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 12278.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1406999111/-/DCSupplemental.
References
- 1.Cummings M, Breitling R, Takano E. Steps towards the synthetic biology of polyketide biosynthesis. FEMS Microbiol Lett. 2014;351(2):116–125. doi: 10.1111/1574-6968.12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford JM, Townsend CA. New insights into the formation of fungal aromatic polyketides. Nat Rev Microbiol. 2010;8(12):879–889. doi: 10.1038/nrmicro2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chooi YH, Tang Y. Navigating the fungal polyketide chemical space: From genes to molecules. J Org Chem. 2012;77(22):9933–9953. doi: 10.1021/jo301592k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kirschning A, Hahn F. Merging chemical synthesis and biosynthesis: A new chapter in the total synthesis of natural products and natural product libraries. Angew Chem Int Ed Engl. 2012;51(17):4012–4022. doi: 10.1002/anie.201107386. [DOI] [PubMed] [Google Scholar]
- 5.Kwon SJ, Mora-Pale M, Lee MY, Dordick JS. Expanding nature’s small molecule diversity via in vitro biosynthetic pathway engineering. Curr Opin Chem Biol. 2012;16(1-2):186–195. doi: 10.1016/j.cbpa.2012.02.001. [DOI] [PubMed] [Google Scholar]
- 6.Winter JM, Tang Y. Synthetic biological approaches to natural product biosynthesis. Curr Opin Biotechnol. 2012;23(5):736–743. doi: 10.1016/j.copbio.2011.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thomas GL, Johannes CW. Natural product-like synthetic libraries. Curr Opin Chem Biol. 2011;15(4):516–522. doi: 10.1016/j.cbpa.2011.05.022. [DOI] [PubMed] [Google Scholar]
- 8.Xu J, Jiang CS, Zhang ZL, Ma WQ, Guo YW. Recent progress regarding the bioactivities, biosynthesis and synthesis of naturally occurring resorcinolic macrolides. Acta Pharmacol Sin. 2014;35(3):316–330. doi: 10.1038/aps.2013.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Winssinger N, Fontaine JG, Barluenga S. Hsp90 inhibition with resorcyclic acid lactones (RALs) Curr Top Med Chem. 2009;9(15):1419–1435. doi: 10.2174/156802609789895665. [DOI] [PubMed] [Google Scholar]
- 10.Wei H, et al. Monocillin II inhibits human breast cancer growth partially by inhibiting MAPK pathways and CDK2 Thr160 phosphorylation. ChemBioChem. 2012;13(3):465–475. doi: 10.1002/cbic.201100558. [DOI] [PubMed] [Google Scholar]
- 11.Gaffoor I, Trail F. Characterization of two polyketide synthase genes involved in zearalenone biosynthesis in Gibberella zeae. Appl Environ Microbiol. 2006;72(3):1793–1799. doi: 10.1128/AEM.72.3.1793-1799.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang S, et al. Functional characterization of the biosynthesis of radicicol, an Hsp90 inhibitor resorcylic acid lactone from Chaetomium chiversii. Chem Biol. 2008;15(12):1328–1338. doi: 10.1016/j.chembiol.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 13.Reeves CD, Hu Z, Reid R, Kealey JT. Genes for the biosynthesis of the fungal polyketides hypothemycin from Hypomyces subiculosus and radicicol from Pochonia chlamydosporia. Appl Environ Microbiol. 2008;74(16):5121–5129. doi: 10.1128/AEM.00478-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu Y, et al. Characterization of the biosynthetic genes for 10,11-dehydrocurvularin, a heat shock response-modulating anticancer fungal polyketide from Aspergillus terreus. Appl Environ Microbiol. 2013;79(6):2038–2047. doi: 10.1128/AEM.03334-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foulke-Abel J, Townsend CA. Demonstration of starter unit interprotein transfer from a fatty acid synthase to a multidomain, nonreducing polyketide synthase. ChemBioChem. 2012;13(13):1880–1884. doi: 10.1002/cbic.201200267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford JM, et al. Structural basis for biosynthetic programming of fungal aromatic polyketide cyclization. Nature. 2009;461(7267):1139–1143. doi: 10.1038/nature08475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, et al. Classification, prediction, and verification of the regioselectivity of fungal polyketide synthase product template domains. J Biol Chem. 2010;285(30):22764–22773. doi: 10.1074/jbc.M110.128504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu Y, et al. Rational reprogramming of fungal polyketide first-ring cyclization. Proc Natl Acad Sci USA. 2013;110(14):5398–5403. doi: 10.1073/pnas.1301201110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang M, Zhou H, Wirz M, Tang Y, Boddy CN. A thioesterase from an iterative fungal polyketide synthase shows macrocyclization and cross coupling activity and may play a role in controlling iterative cycling through product offloading. Biochemistry. 2009;48(27):6288–6290. doi: 10.1021/bi9009049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu Y, et al. Thioesterase domains of fungal nonreducing polyketide synthases act as decision gates during combinatorial biosynthesis. J Am Chem Soc. 2013;135(29):10783–10791. doi: 10.1021/ja4041362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vagstad AL, et al. Combinatorial domain swaps provide insights into the rules of fungal polyketide synthase programming and the rational synthesis of non-native aromatic products. Angew Chem Int Ed Engl. 2013;52(6):1718–1721. doi: 10.1002/anie.201208550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Korman TP, et al. Structure and function of an iterative polyketide synthase thioesterase domain catalyzing Claisen cyclization in aflatoxin biosynthesis. Proc Natl Acad Sci USA. 2010;107(14):6246–6251. doi: 10.1073/pnas.0913531107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou H, et al. Enzymatic synthesis of resorcylic acid lactones by cooperation of fungal iterative polyketide synthases involved in hypothemycin biosynthesis. J Am Chem Soc. 2010;132(13):4530–4531. doi: 10.1021/ja100060k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou H, Qiao K, Gao Z, Vederas JC, Tang Y. Insights into radicicol biosynthesis via heterologous synthesis of intermediates and analogs. J Biol Chem. 2010;285(53):41412–41421. doi: 10.1074/jbc.M110.183574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu Y, et al. Insights into the biosynthesis of 12-membered resorcylic acid lactones from heterologous production in Saccharomyces cerevisiae. ACS Chem Biol. 2014;9(5):1119–1127. doi: 10.1021/cb500043g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vagstad AL, Hill EA, Labonte JW, Townsend CA. Characterization of a fungal thioesterase having Claisen cyclase and deacetylase activities in melanin biosynthesis. Chem Biol. 2012;19(12):1525–1534. doi: 10.1016/j.chembiol.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou H, et al. A fungal ketoreductase domain that displays substrate-dependent stereospecificity. Nat Chem Biol. 2012;8(4):331–333. doi: 10.1038/nchembio.912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McDaniel R, et al. Multiple genetic modifications of the erythromycin polyketide synthase to produce a library of novel “unnatural” natural products. Proc Natl Acad Sci USA. 1999;96(5):1846–1851. doi: 10.1073/pnas.96.5.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wong FT, Khosla C. Combinatorial biosynthesis of polyketides—A perspective. Curr Opin Chem Biol. 2012;16(1-2):117–123. doi: 10.1016/j.cbpa.2012.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu MC, Law B, Wilkinson B, Micklefield J. Bioengineering natural product biosynthetic pathways for therapeutic applications. Curr Opin Biotechnol. 2012;23(6):931–940. doi: 10.1016/j.copbio.2012.03.008. [DOI] [PubMed] [Google Scholar]
- 31.Menzella HG, Carney JR, Santi DV. Rational design and assembly of synthetic trimodular polyketide synthases. Chem Biol. 2007;14(2):143–151. doi: 10.1016/j.chembiol.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 32.Kodadek T. The rise, fall and reinvention of combinatorial chemistry. Chem Commun (Camb) 2011;47(35):9757–9763. doi: 10.1039/c1cc12102b. [DOI] [PubMed] [Google Scholar]
- 33.Zhou H, Zhan J, Watanabe K, Xie X, Tang Y. A polyketide macrolactone synthase from the filamentous fungus Gibberella zeae. Proc Natl Acad Sci USA. 2008;105(17):6249–6254. doi: 10.1073/pnas.0800657105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu T, Chiang YM, Somoza AD, Oakley BR, Wang CC. Engineering of an “unnatural” natural product by swapping polyketide synthase domains in Aspergillus nidulans. J Am Chem Soc. 2011;133(34):13314–13316. doi: 10.1021/ja205780g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wijeratne EM, et al. Geopyxins A-E, ent-kaurane diterpenoids from endolichenic fungal strains Geopyxis aff. majalis and Geopyxis sp. AZ0066: Structure-activity relationships of geopyxins and their analogues. J Nat Prod. 2012;75(3):361–369. doi: 10.1021/np200769q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Santagata S, et al. Using the heat-shock response to discover anticancer compounds that target protein homeostasis. ACS Chem Biol. 2012;7(2):340–349. doi: 10.1021/cb200353m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLellan CA, et al. A rhizosphere fungus enhances Arabidopsis thermotolerance through production of an HSP90 inhibitor. Plant Physiol. 2007;145(1):174–182. doi: 10.1104/pp.107.101808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Busch B, et al. Multifactorial control of iteration events in a modular polyketide assembly line. Angew Chem Int Ed Engl. 2013;52(20):5285–5289. doi: 10.1002/anie.201301322. [DOI] [PubMed] [Google Scholar]
- 39.Liu T, Sanchez JF, Chiang YM, Oakley BR, Wang CC. Rational domain swaps reveal insights about chain length control by ketosynthase domains in fungal nonreducing polyketide synthases. Org Lett. 2014;16(6):1676–1679. doi: 10.1021/ol5003384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma SM, et al. Complete reconstitution of a highly reducing iterative polyketide synthase. Science. 2009;326(5952):589–592. doi: 10.1126/science.1175602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee KK, Da Silva NA, Kealey JT. Determination of the extent of phosphopantetheinylation of polyketide synthases expressed in Escherichia coli and Saccharomyces cerevisiae. Anal Biochem. 2009;394(1):75–80. doi: 10.1016/j.ab.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 42.Turbyville TJ, et al. Search for Hsp90 inhibitors with potential anticancer activity: Isolation and SAR studies of radicicol and monocillin I from two plant-associated fungi of the Sonoran desert. J Nat Prod. 2006;69(2):178–184. doi: 10.1021/np058095b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.