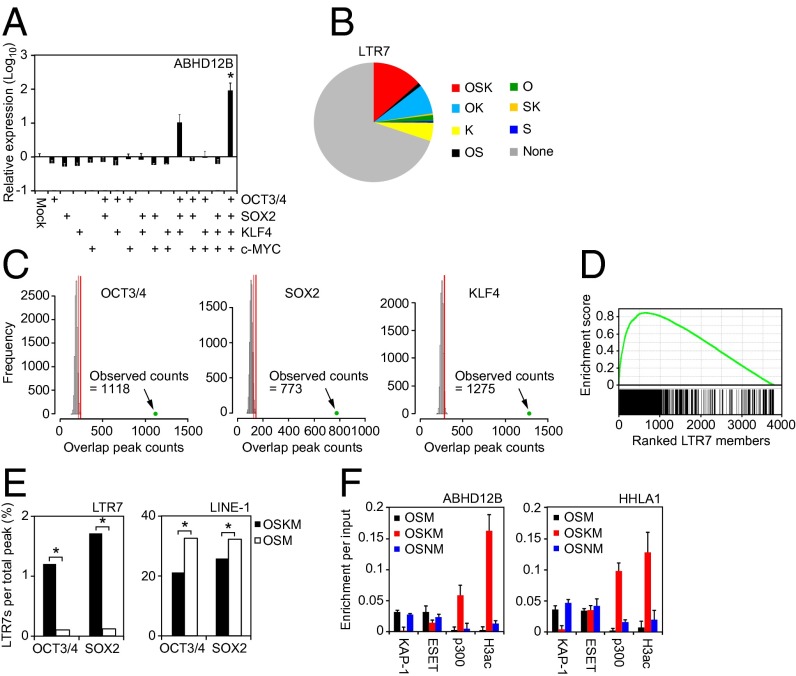
Fig. 4.
Role of OSK in LTR7 activation. (A) OSK is required for activation of ABHD12B expression. Relative expression level of ABHD12B on day 7 posttransduction for all combinations of OSKM. Error bars are SDs. n = 3. *P < 0.05 vs. Mock was calculated by Dunnett test. (B) Distribution of reprogramming factor occupancy on all LTR7s loci revealed by ChIP-seq. (C) Significance of the interaction of reprogramming factors to LTR7s. Histograms show counts of peaks for OCT3/4, SOX2, or KLF4 overlapped with randomly selected regions (10,000 random trials). The 95th percentile count of distribution is marked by red lines. Green dots show counts of ChIP-seq peaks on LTR7 regions with overhang sequences (250 bp). (D) GSEA plot showing enrichment of OSK occupancies in expressed LTR7s. Expressed LTR7 family members in TRA-1-60 (+) cells on day 15 are enriched in the set of LTR7s that show full-array OSK binding (P = 2.4e-155). (E) KLF4-dependent binding of OCT3/4 and SOX2 to LTR7s. Bars show the percentage of OCT3/4- or SOX2-bound LTR7 family members and LINE-1 in HDFs transduced with OSKM (closed) or OSM (open) on day 3 posttransduction. χ2 tests were performed between the proportions (*P < 0.05). (F) Interaction between HERV-H loci and chromatin modifiers. ChIP assays were performed to analyze the interaction of ABHD12B and HHLA1 loci with KAP-1, ESET, p300, and pan-acetyl histone H3 (H3ac) occupancy in HDFs transduced with OSM, OSKM, or OSNM on day 3 were analyzed by ChIP-qPCR. n = 3. Error bars are SD.