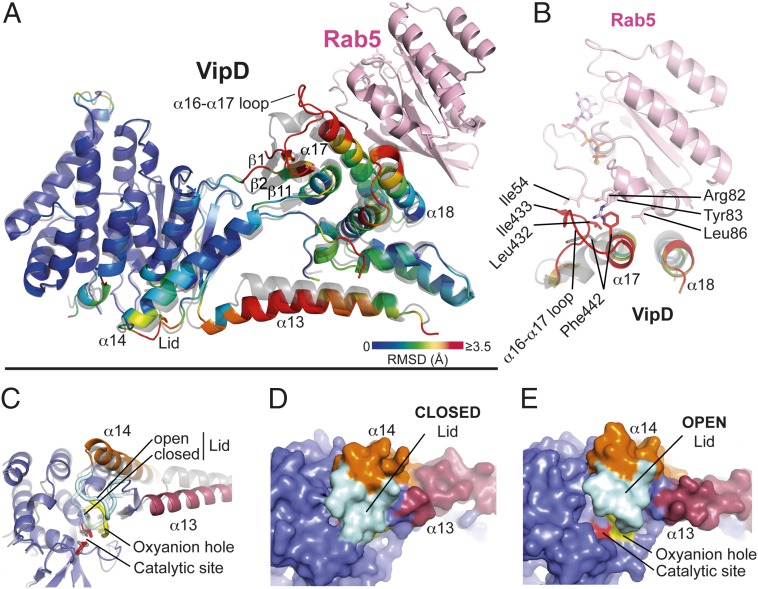
Fig. 2.
Allosteric activation of VipD through Rab5 binding. (A) Structural changes in VipD on Rab5 binding. Rab5c18–182 (colored in pink) is complexed to VipD19–564, which is colored from slate to red based on the root mean square deviation (RMSD) of C-α atom pairs when superimposed with the unbound form of VipD19–564 (PDB ID code 4AKF) shown in transparent gray. The black line represents the membrane plane. (B) Close-up of VipD–Rab5 interaction. The α17-α18 loop of VipD undergoes a Rab5-induced conformational rearrangement resulting in residue Phe442 of VipD being inserted into a hydrophobic pocket formed by Arg82, Tyr83, and Leu86 of Rab5. The displacement of the α16–17 loop favors the hydrophobic interaction between Leu432 and Ile433 of VipD with Ile54 of Rab5. Color code as in A. The remaining VipD structure has been omitted for clarity. (C) Close-up view of the catalytic site highlighting displacement of the lid (β10-α14 loop, light blue). (D and E) Surface representation of the unbound (D) and Rab5-bound (E) VipD molecule, respectively. Same view as in C.