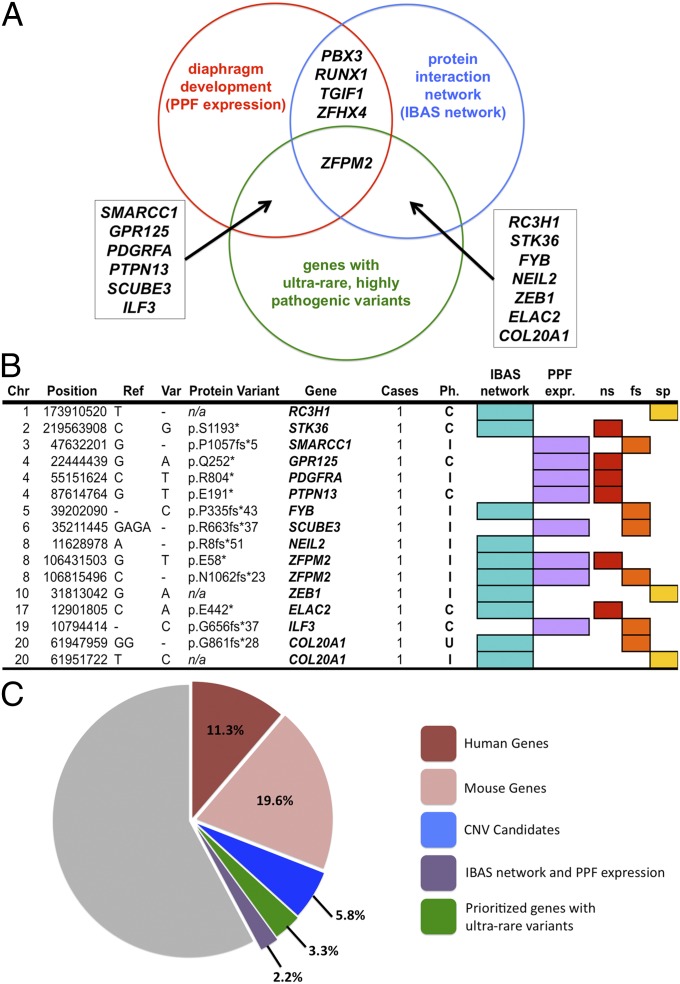
Fig. 2.
Candidate genes. (A) Candidate genes derived from the overlap of PPF transcriptomes, IBAS protein interaction networks, and genes with ultrarare and highly deleterious SNVs. (B) Ultrarare and highly deleterious SNVs in prioritized genes. C, complex CDH; Chr, chromosome; fs, frameshift; I, isolated CDH; n/a, not applicable; ns, nonsense; Ph., phenotype; Ref., reference allele; sp, splice site loss; Var, variant allele. (C) Exomes (%) with predicted pathogenic SNVs in human (red, 11.3%) or mouse (pink, 19.6%) genes, CNV candidates (blue, value = 5.8%), candidates selected by PPF transcriptomes and IBAS networks (green, value = 3.3%), and in genes with ultrarare and highly disruptive variants (2.2%).