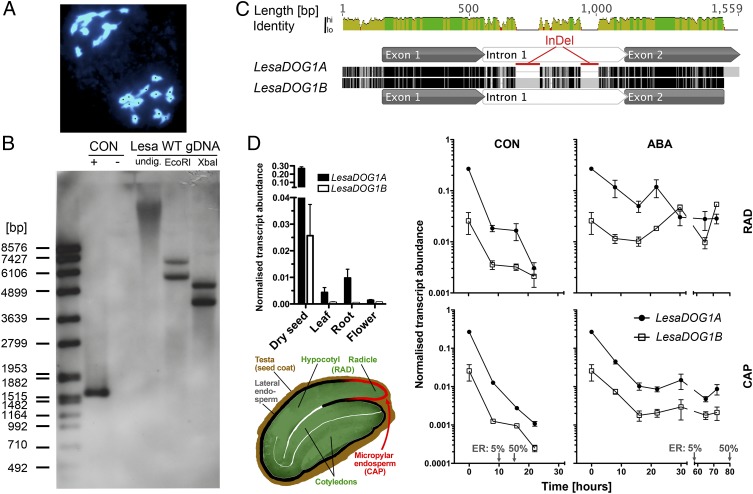
Fig. 1.
Two seed-expressed DOG1 genes, LesaDOG1A and LesaDOG1B, in the diploid (2n = 24) species L. sativum. (A) DAPI-stained meiotic (metaphase II) chromosome spreads from flower bud tissue of L. sativum exhibiting 12 chromosomes, thus demonstrating regular meiosis. For better counting, chromosomes are labeled with black dots. (B) Southern blot analysis of the L. sativum FR14 genome indicates the presence of two DOG1 genes. Genomic DNA (Lesa WT gDNA) undigested (undig.) or digested with EcoRI or XbaI was hybridized with a LesaDOG1A probe. Hybridization controls were plasmids with (+) or without (−) LesaDOG1A full-length gDNA inserts. The left lane shows the DNA molecular mass ladder. CON, control. (C) Pairwise alignment of LesaDOG1A and LesaDOG1B (nearly full-length) gDNA sequences. Identical residues are colored black; mismatches are colored light gray; gaps are indicated by horizontal lines. Exon-intron annotations were derived by comparison with the respective cDNAs. Two large intronic InDels are marked in red. (D) Transcript abundances (Upper Left) of LesaDOG1A and LesaDOG1B in different plant tissues and during germination in the seed RAD and CAP tissues (as indicated by the schematic L. sativum seed drawing, Lower Left) determined by qRT-PCR analysis. n = 3; data are shown as mean ± SEM. (Right) Seeds were imbibed without (CON) or with the addition of 10 µM ABA. Gray arrows indicate time points of 5% and 50% ER. RAD comprises the radicle plus ca. one-third of the lower hypocotyl (the embryo growth zone); CAP is the micropylar endosperm tissue. Note that the dry seed samples (0 h) of the qRT-PCR analysis represent CAP+RAD tissue and not whole seeds.