Fig. 6.
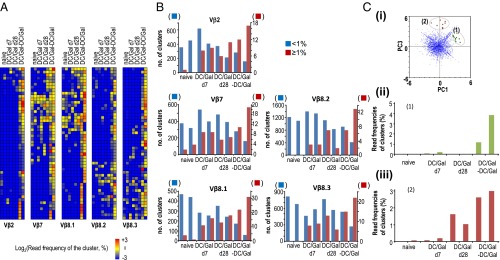
TCR repertoire analyses of KLRG1+ iNKT cells. The naive iNKT cells and KLRG1+ iNKT cells were sorted and tested for TCR RNA-seq analysis. In brief, pooled naive or KLRG1+ iNKT cells were prepared from five or six naive or DC/Gal-immunized mice on day 7 or day 28. In some experiments, DC/Gal-injected mice were boosted with DC/Gal 28 d after the first immunization (DC/Gal-DC/Gal). The CDR3β of naive iNKT cells or KLRG1+ iNKT cells of DC/Gal-injected mice (at day 7 or day 28) or DC/Gal-DC/Gal–boosted mice was assessed by a GS Junior system (Roche) in two independent experiments at each stage. (A) The clusters of high-frequency (top50) CDR3 in each Vβ repertoire in the boosted mice were selected and then clustered among those in naive mice, DC/Gal-injected mice (at day 7 or day 28), or DC/Gal-DC/Gal–boosted mice. The results are shown as heatmaps indicating the cluster sizes [log2(frequency of the sequence reads in the cluster) (%)]. (B) The size distributions of CDR3β clusters in each Vβ repertoire of each group of mice. The numbers of clusters that are larger or smaller than 1% of the total sequence reads are shown by red and blue bars, respectively. (C) (i) Principal component analysis was performed for whole clusters of CDR3. Two groups of CDR3 clusters were identified as groups 1 and 2. (ii and ii) The cluster size distributions of KLRG1+ iNKT cells in groups 1 and 2 are shown as bar graphs in ii and iii, respectively.
