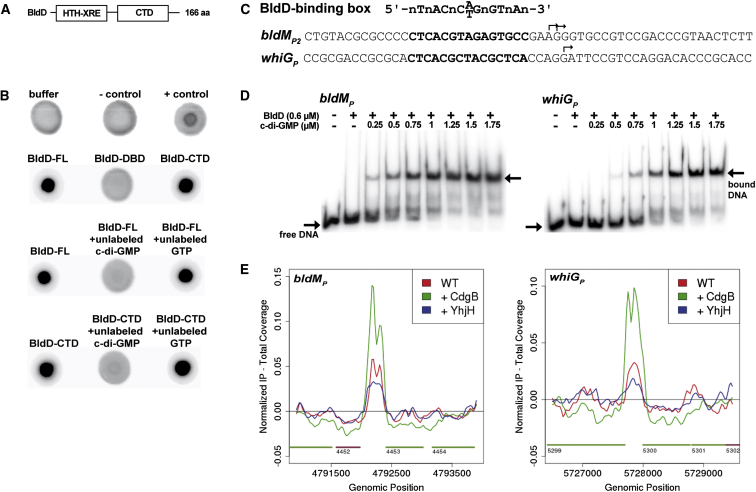
Figure 2.
The BldD CTD Binds c-di-GMP and Activates DNA Binding
(A) Schematic showing the domain organization of the BldD protein. BldD contains an N-terminal DBD connected by a flexible linker to a CTD.
(B) Results of DRaCALAs assays carried out using purified FL His6-BldD, His6-BldD-DBD or His6-BldD-CTD, and 32P-labeled c-di-GMP. The TetR-like regulator SVEN_1547 and the active DGC PleD∗ from C. crescentus served as negative (−) and positive (+) controls, respectively (top row). In competition DRaCALAs, excess cold c-di-GMP or GTP was added to the binding reaction containing 32P-labeled c-di-GMP and His6-FL BldD or His6-BldD-CTD. c-di-GMP binding is indicated by dark spots centered on the nitrocellulose.
(C) Top: the DNA consensus motif bound by BldD (den Hengst et al., 2010). Below are the sequences from the S. venezuelae bldM and whiG promoter regions (with BldD-binding boxes in bold). The transcriptional start sites are indicated by bent arrows.
(D) EMSA analyses of BldD binding to the bldM and whiG promoters ± c-di-GMP. Free DNA and protein-DNA complexes (bound DNA) are indicated with arrows.
(E) In vivo BldD ChIP-seq data for bldM and whiG. Color coding of the ChIP samples is as follows: WT S. venezuelae (red), S. venezuelae overexpressing the S. coelicolor DGC CdgB (green), and S. venezuelae overexpressing the E. coli PDE YhjH (blue). Plots span ∼3 kb of DNA sequence. Genes running left to right are shown in green, and genes running right to left are shown in red.