Figure S6.
Structural Analyses of the BldD DBD and the Full-Length BldD-(c-di-GMP)-DNA Complex, Related to Figure 7
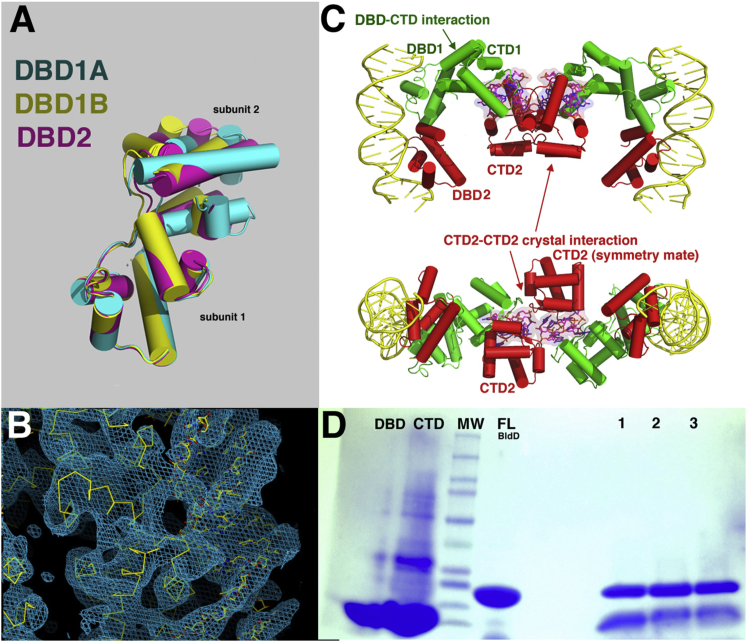
(A) Overlay of one subunit of the crystallographic dimer of the S. coelicolor BldD DBD (yellow) and one subunit of the two crystallographic dimers observed in the S. venezuelae DBD structure (magenta and cyan). This overlay shows that the intersubunit interfaces of these dimers are small and likely not physiologically relevant (∼300 Å2 BSA buried) and are not the same.
(B) An electron density map calculated with phases from the 4.5 Å resolution structure of the BldD-(c-di-GMP)-21-mer complex and contoured at 1σ.
(C) Two views of the crystal structure of the S. venezuelae FL BldD-(c-di-GMP)-DNA complex. One BldD subunit is red and the other green. The c-di-GMP molecules are shown as sticks and surfaces and colored magenta. The DNA is shown as a yellow cartoon. The DNA forms a pseudocontinuous helix in the crystal, which stabilizes the DBD-DNA interaction while the c-di-GMP dimerized CTDs are flexibly tethered to their DBDs. One of the CTDs (green) makes a weak interaction with its DBD while the other CTD (red) is fastened in place in the crystal via contacts to a symmetry-related CTD.
(D) Proteolysis of FL S. venezuelae BldD by endoprotease Glu-C. Lane 1: BldD-(c-di-GMP)-DNA, Lane 2: BldD-DNA, Lane 3: apo FL BldD. Neither the presence of DNA nor c-di-GMP protects the linker region between the DBD and CTD from proteolysis.