Fig. 1.
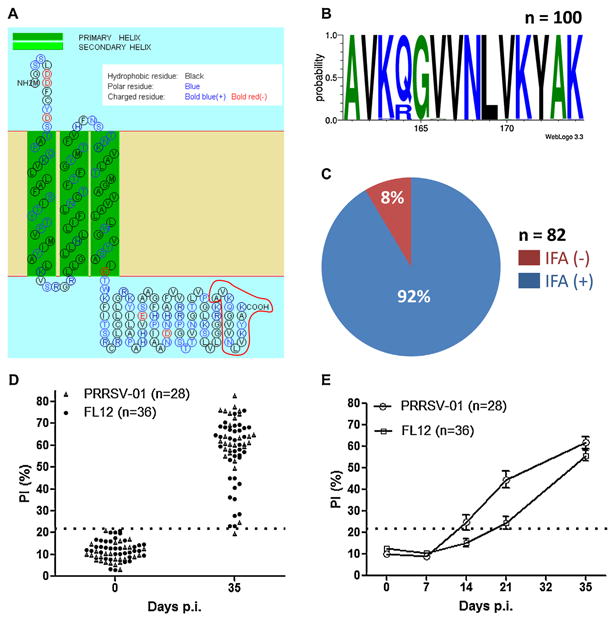
Epitope-M201 is highly conserved and immunodominant. (A) Prediction of transmembrane helixes and topology of the PRRSV strain FL12 M protein. The figure was generated through the use of the web based application SOSUI engine ver. 1.11. Epitope-M201 is outlined in red at the carboxyl-terminus of the protein. (B) Sequence logo of epitope-M201 generated from 100 type-II PRRSV M protein sequences collected from GenBank. The relative positions of the amino acids are indicated in the X-axis. The height of symbols within the stack indicates the relative frequency of each amino at that position. The figure was generated through the use of the web based application WebLogo 3.3. (C) Reactivity of MAb-201 with 82 type-II PRRSV strains/isolates as determined by indirect immunofluorescence assay (IFA). (D) Validation of the bELISA. The positive and negative control sera were collected from 64 pigs before infection and at 35 days p.i. (details of the serum samples are described in Section 2). (E) Kinetics of seroconversion to epitope-M201 as determined by bELISA. The sera were collected from the same pigs mentioned in panel (D). The horizontal dotted line represents the cut-off of the assay. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
