Abstract
Genetic differentiation can be promoted allopatrically by geographic isolation of populations due to limited dispersal ability and diversification over time or sympatrically through, for example, host-race formation. In crop pests, the trading of crops across the world can lead to intermixing of genetically distinct pest populations. However, our understanding of the importance of allopatric and sympatric genetic differentiation in the face of anthropogenic genetic intermixing is limited. Here, we examined global sequence variation in two mitochondrial and one nuclear genes in the seed beetle Callosobruchus maculatus that uses different legumes as hosts. We analyzed 180 samples from 42 populations of this stored bean pest from tropical and subtropical continents and archipelagos: Africa, the Middle East, South and Southeast Asia, Oceania and South America. For the mitochondrial genes, there was weak but significant genetic differentiation across continents/archipelagos. Further, we found pronounced differentiation among subregions within continents/archipelagos both globally and within Africa but not within Asia. We suggest that multiple introductions into Asia and subsequent intermixing within Asia have generated this pattern. The isolation by distance hypothesis was supported globally (with or without continents controlled) but not when host species was restricted to cowpeas Vigna unguiculata, the ancestral host of C. maculatus. We also document significant among-host differentiation both globally and within Asia, but not within Africa. We failed to reject a scenario of a constant population size in the recent past combined with selective neutrality for the mitochondrial genes. We conclude that mitochondrial DNA differentiation is primarily due to geographic isolation within Africa and to multiple invasions by different alleles, followed by host shifts, within Asia. The weak inter-continental differentiation is most likely due to frequent inter-continental gene flow mediated by human crop trade.
Introduction
Genetic differentiation can be induced allopatrically by geographic isolation of populations and/or sympatrically by, for example, host-race formation. In crop pests, the trade of crops across the world tends to generate a global mixing of genetic variants in pest populations, whether these were generated and maintained either allopatrically (e.g., [1],[2]) or sympatrically (e.g., [3],[4]). However, the relative contributions of allopatric or sympatric genetic differentiation versus global genotypic intermixing by natural [5] or human-aided [6]–[8] dispersal to global (continental and island) population structures has rarely been evaluated quantitatively (but see [9],[10]).
Association with specific host plants may allow the formation of genetically distinct pest host races in some cases [6], but not in others [5]. It has been suggested that an increased use of dry crops may have led to the multivoltine life cycles, polyphagy and expanded geographic distribution of stored bean pests [11]–[16]. Among the stored bean pests are several seed beetle species of the genus Callosobruchus (Coleoptera: Chrysomelidae: Bruchinae). Bruchine adults of such species lay eggs on dry seeds of legume crops and larvae burrow into them. Low levels of infestation are difficult to detect because these beetles live inside the seeds as larvae, pupae and pre-emergence adults. Because of the cryptic feeding in grains, world trade of dry beans for human consumption can potentially lead to the effective spread of these beetles.
In general, flight distance is correlated with body size in animals [17]. Among insects, beetles are generally weak flyers. For small beetles like seed beetles, the flight distance is limited [18] and long-distance transoceanic dispersal, even via air currents, is considered virtually impossible [19]–[21]. The low water content of stored beans is known to increase the frequency of the flightless (sedentary) form in Callosobruchus populations [22],[23], thus further limiting the dispersal of populations of these beetles.
Callosobruchus maculatus (F.) is a major stored bean pest in sub-Saharan Africa [24],[25]. It is a model organism for studies of behavior, polymorphism, population ecology, and evolutionary biology (e.g., [26]–[44]). It is considered to be of Afrotropical origin [45]. Its main, and probably ancestral, host is the cowpea Vigna unguiculata (L.) Walp., but other species of Vigna and other cultivated legumes are also infested by the beetle [16],[45]. Recent molecular work has revealed that C. maculatus populations are partially intermixed across agro-ecological zones of Western Africa [46].
The cowpea is a drought-tolerant food crop for humans and livestock feed that is grown throughout the semi-arid tropics [47]–[49]. It originated in southeast Africa and expanded its distribution westward and southward [50],[51]. Its domestication is estimated to have taken place in Western Africa based on maximum diversity of cultivated cowpeas [50]–[52]. It has been estimated that it was brought into India about 2,200 years ago and it has been cultivated widely in China, India and Southeast Asia [49] since. In the 17th century it was introduced to the tropical America by the Spaniards and is now widely cultivated in the United States, Caribbean islands and Brazil [49]. Callosobruchus maculatus also uses other economically important bean species as hosts, such as V. radiata and V. angularis [35],[37],[53], Vicia faba, Cicer arietinum and Cajanus cajan [16],[45],[54],[55]. Although less important as a crop, the bambara groundnut V. subterranea of Afrotropical origin [51] is also attacked by the beetle.
Here, we aimed to study the effects of geographic isolation by limited dispersal ability and genetic differentiation over time, host-plant utilization by formation of distinct host-associated races, and anthropogenic gene flow by trade of crop on the global genetic differentiation of C. maculatus, based on multiple mitochondrial and nuclear gene sequences of 42 populations that use different bean species as hosts. We evaluated these factors by comparing genetic variation between hierarchical spatial scales (continents, subregions and local populations) and between populations using different host plants. Human-aided dispersal is expected to lead to weak or no differentiation at the spatial scale where anthropogenically mediated gene flow is most frequent. We tested for congruence between the C. maculatus phylogeny and geography on one hand and between the phylogeny and host plant usage on the other. Furthermore, nucleotide sequence variation was compared among populations across five different continents and among those using different host plant species. The isolation-by-distance hypothesis was tested globally and within continents and with host plant controlled.
Materials and Methods
Ethics statement
No specific permits were required for the described field studies. No specific permission was required for sampling at any locations and activity. The locations are not privately owned or protected in any way. No activity during the field study involved endangered species or protected species.
Study animals
Callosobruchus maculatus were collected as adults that emerged from randomly selected cultivated beans collected from local markets in various parts of Africa, the Middle East, South and Southeast Asia, Oceania and South America (Table 1). Populations collected in the mid-1990s or earlier were reared in the laboratory for various period of time (see Table 1). The beetles were preserved in acetone until used for DNA extraction. We used 180 sequences for phylogenetic inference and congruence and 160 sequences of recent sampling for other analyses (see the following sections).
Table 1. Collection data for the Callosobruchus maculatus populations.
| Region | Subregion | Code | n c | Host species (n c) |
| Population | ||||
| Africa | Western Africa | |||
| Malib | Mal | 4 | NA | |
| Tamale, Ghanab | Gh | 2 | NAa | |
| Beninb | Ben | 2 | NA | |
| Lossa, Burkina Fasob | LosBF | 5 | NA | |
| Oyo, Nigeriab | OyNg | 5 | NA | |
| Borno, Nigeria | BoNg | 3 | Vigna unguiculata | |
| northern Nigeria | nNg | 7 | Vigna unguiculata | |
| Lomé, Togo | LomTg | 6 | Vigna unguiculata | |
| Nlongkak, Cameroon | Cam | 3 | Vigna unguiculata | |
| Democratic Republic of the Congo (Zaire)b | Zai | 2 | NA | |
| Central Africa | ||||
| Uganda | Ug | 3 | NA | |
| Madagascar | ||||
| Manjakandriana, Madagascar | MaMd | 9 | Vigna unguiculata | |
| Antananarivo, Madagascar | AnMd | 9 | Vigna subterranea | |
| Moramanga, Madagascar | MoMd | 3 | Vigna subterranea | |
| Middle East | Middle East | |||
| Lattakia, Syria | Sy | 2 | Pisum sativum (1), Cicer arietinum (1) | |
| Oman | Om | 3 | NA | |
| Asia | India/Sri Lanka/Nepal | |||
| Burdwan, India | BuIn | 8 | Cajanus cajan | |
| Columbo, Sri Lanka | SL | 5 | Vigna unguiculata (4), NA (1) | |
| Kathmandu, Nepal | Np | 9 | Vigna unguiculata | |
| Thailand/Myanmar | ||||
| Yangon, Myanmar | My | 2 | Vigna unguiculata | |
| Bangkok, Thailand | BanTh | 7 | Vigna unguiculata | |
| Chom Thong, Thailand | CTTh | 2 | Vigna angularis | |
| Chiang Mai, Thailand | CMTh | 6 | Vigna radiata (3), Vigna angularis (3) | |
| China/Vietnam | ||||
| Kunming, Yunnan, China | KmCh | 8 | Vigna unguiculata | |
| Hainan, China | HaiCh | 8 | Vigna radiata | |
| Thanh Hoa, Vietnam | THVi | 5 | Vigna unguiculata (4), Vigna radiata (1) | |
| Tam Ky, Vietnam | TKVi | 2 | Vigna unguiculata | |
| Taiwan/Japan | ||||
| Yungkang, Tainan, Taiwan | TnTw | 7 | Vigna unguiculata (4), Vigna radiata (3) | |
| Taipei, Taiwan | TpTw | 3 | Vigna radiata (1), Vigna angularis (2) | |
| Okinawa, Japan | OkiJ | 1 | Vigna unguiculata | |
| Indonesia/Philippines | ||||
| Los Banos, Laguna, Philippines | Ph | 8 | Vigna radiata | |
| Bali, Indonesia | Bal | 8 | Vigna radiata | |
| Java, Indonesia | Jav | 2 | Vigna radiata | |
| Kota Kinabalu, Malaysia | KK | 9 | Vigna radiata | |
| Oceania | New Zealand | |||
| New Zealand | NZ | 3 | Vicia faba | |
| South America | Brazil | |||
| Uberlandia, Brazil | Br | 9 | Vigna unguiculata |
Locations are categorized by regions (continents or archipelagos) and by subregions.
excluded from dataset for AMOVA, IBD test and sudden demographic growth test because of >10 years of laboratory rearing and/or small sample sizes.
number of individuals used.
DNA sequencing
Whole genomic DNA was extracted from individual beetles using a DNeasy Blood & Tissue Kit (QIAgen, Tokyo, Japan). We amplified and sequenced two mitochondrial fragments, COI and tRNALeu–COII, and nuclear EF-1α (elongation factor-1α) and two fragments from 28S rRNA. For the PCR and sequencing, we followed the protocol previously described in [6],[14]. The primers used were COI-F (5′-ATAATTTTTTTTATAGTTATACC-3′) and COI2-2 [6] for COI, and 5′-TAATATGGCAGATTAGTGCATTGGA-3′ and 5′-GAGACCATTACTTGCTTTCAGTCATCT-3′ ([56], modified from [57]; see [14] for details) for tRNALeu–COII. Nuclear EF-1α fragment was amplified using primers 5′-GGTATCACCATTGATATTGCHTTDTGGAA-3′ and 5′-ACCAGCAACATAACCACGACG-3′ [58]. Two nuclear 28S rRNA gene fragments were amplified using the primers, 28S-01 (5′-GACTACCCCCTGAATTTAAGCAT-3′) in combination with 28S-R01 (5′-GACTCCTTGGTCCGTGTTTCAAG-3′) for D2–D3 fragment [59] and 28SD4-5-F in combination with 28SD4-5-R for D4–D5 fragment [60]. The PCRs were performed following [6] with a total volume of 15 µl; initial preheating at 95°C for 2 min, followed by 35–40 cycles of a denaturation phase at 94°C for 30 s, an annealing at 45°C for COI, 51°C for tRNALeu–COII, a constant temperature between 44–52°C for EF-1α, 54°C for 28S-01 and 51°C for 28SD for 40 s, and an extension at 60°C for the mitochondrial genes, 62°C for the EF-1α and at 70°C for the 28S fragments, for 1 min–1 min 10 s. Sequencing was done with BigDye Terminator v3.1 Cycle Sequencing Kit (Life Technologies/Applied Biosystems) on 3730 DNA Analyzer (Applied Biosystems).
Because infection by and vertical transmission of the intracellular symbiont Wolbachia may select for mitochondrial haplotypes in infected females through cytoplasmic incompatibility [61], we used PCR to diagnose possible Wolbachia infection using primers fts-Z-f and fts-Z-r for the ftsZ gene coding fragment [62]. Here, beetles of Callosobruchus chinensis (L.) that were naturally infected with Wolbachia were used as a positive control [63].
Reconstruction of molecular phylogeny
The phylogenetic relationships among the beetle populations were estimated by Bayesian inference using MrBayes 3.2 [64],[65]. Callosobruchus imitator Kingsolver (Genbank accessions DQ459044, DQ459028) and C. subinnotatus (Pic) (DQ459047, DQ459031) were used as outgroups (see [14]). An evolutionary model for reconstruction of the molecular phylogeny for the populations was selected from the 24 models in MrBayes 3.2 [65] using MrAIC.pl 1.3.1 [66]. For independent gene trees, the HKY+Γ model (Hasegawa, Kishino and Yano model [67], with a gamma distribution of rates across sites) was supported both for COI and for tRNALeu–COII by AICc. For all model parameters, we used the default priors. The mitochondrial gene sequence data were subjected to two independent parallel runs each with four Metropolis-coupled chains [68] of which three were heated (i.e., accepted at a higher rate than specified by the Metropolis-Hastings criterion) using MCMC (Markov chain Monte Carlo, [69],[70]) algorithms, sampled every 1,000 generations. The convergence of parameters among runs was checked using Tracer 1.5.0 [71]. The separate gene trees were estimated using similar parameters with 6 million generation runs. The combined data set was subjected to 10 million generation runs with a partition between COI and tRNALeu–COII. The first 25% of the total number of generations were discarded as a burn-in phase. For comparison of congruence of phylogenetic topology with geography and host plants (see the next section), we used a subset of the data with host plant information (Table 1) and re-estimated the C. maculatus phylogeny using MrBayes.
Congruence between C. maculatus phylogeny and geography/hosts
The congruence of the C. maculatus phylogeny with either the geographic subregions (West and Central Africa, Madagascar, the Middle East, India/Sri Lanka/Nepal, Thailand/Myanmar, China/Vietnam, Taiwan/Japan, Indonesia/Philippines, New Zealand, and Brazil) or host plants (V. unguiculata, V. radiata, V. angularis, V. subterranea, Vicieae (Vicia and Pisum), Cajanus, and Cicer) was tested with a topology-dependent permutation tail probability test (T-PTP) implemented in PAUP*4.0 Beta [72] for ingroup taxa only [73]. Randomization of the traits was performed 1,000 times. For this test, we used a Bayesian phylogeny that was estimated from the sequence data after excluding individuals with unknown hosts (see the previous section). We confirmed that this exclusion did not change the basic topology of the phylogeny. Nevertheless, to assure this exclusion would not affect congruence between phylogenetic topology and geography, we also tested the congruence between geography and phylogeny before the exclusion.
Population structure
Here, we used populations of which individuals were analyzed either soon after collection or within a relatively brief rearing history in the lab (160 individuals in total; see Table 1) to minimize any effects of decrease in genetic variability during rearing. Although rearing even for several generations may erode ancestral genetic signature, the laboratory populations we used had been kept at large population sizes to avoid any bottleneck events. We performed hierarchical analyses of molecular variance (AMOVAs) [74]–[76] to compare genetic variances at different hierarchies of either geographic or host plant groups (V. unguiculata, V. angularis, V. radiata, V. subterranea and C. cajan; the other plant species or groups were excluded because of the small sample sizes, Table 1), using Arlequin 3.5.1.3 [77]. Furthermore, to exclude confounding effects resulting from the correlation between geographic regions and host plants, we performed AMOVAs either with geographic regions controlled or with host plants controlled. To control possible effect of unequal sample sizes among populations, we used a resampling procedure where two individuals per population were randomly sampled and subjected to AMOVAs to test statistical robustness of geographic and host-plant effects detected in the whole dataset. We used Tamura and Nei's distance [78]. To correct skewed distribution of nucleotide substitutions among sites, gamma distribution was assumed with α = 0.05 for the mitochondrial genes and α = 0.3 for the nuclear gene, based on our estimation for the C. maculatus dataset.
Isolation by distance
As in the previous analyses, we only used populations that were analyzed soon after collection or within a brief rearing history. We assessed spatial genetic differentiation by testing for isolation by distance (IBD) [79]. With frequent anthropogenic long-distance dispersal events, the IBD correlation is predicted to be weak at most. We tested if, as predicted by IBD, geographic distance (ln-transformed) and F ST/(1- F ST) were positively correlated, by a one-tailed Mantel test based on 2,000 permutations, using the ISOLDE program of Genepop 4.0.10 [80],[81]. We first tested IBD on the global dataset. We then tested for IBD within Africa and within Asia to assess genetic intermixing at a smaller scale. We also restricted this analysis to include only populations using V. unguiculata as a host to exclude possible effect of host-race formation over distance.
Demographic growth and selective neutrality
As in the previous two sections, we used only populations that were subjected to DNA extraction shortly after collection. Recent past sudden demographic expansion was tested in populations on different continents (Africa and Asia) and in populations using the original host V. unguiculata, with raggedness r [82], FS [83] and R 2 [84], using DnaSP5.10.0.1 [85]. Nonsignificant r, significantly negative FS and significantly small R 2 values indicate recent sudden population growth. The latter two indices are statistically more powerful than the raggedness index and FS exhibits high statistical power for large sample sizes, while R 2 is more powerful for small sample sizes [84]. Selective neutrality was tested with Tajima's D [86], using DnaSP. Coalescent simulations were performed for 2,000 iterations to test statistical significance of these indices, using DnaSP.
Results
Molecular phylogeny
The total lengths of the sequenced fragments were 1,726 bp (1,005 bp for COI, 19 bp for tRNALeu, and 702 bp for COII) for the mitochondrial genes (genbank JX297380–JX297419). The sequence data included no indels and the base composition was AT rich, as is typical for insect mitochondrial sequences. The nuclear EF-1α fragment was sequenced for 81 homozygous individuals. We used 866 bp of this fragment, including insertions and deletions. For the nuclear 28S, the sequenced fragments were 1,452 bp in total (822 bp for the D2–D3 fragment and 630 bp for the D4–D5). There was no sequence variation across C. maculatus individuals in the two fragments of 28S and these were thus discarded from subsequent analyses. We analyzed only the mitochondrial trees because homozygous nuclear EF-1α data was available for limited number of individuals.
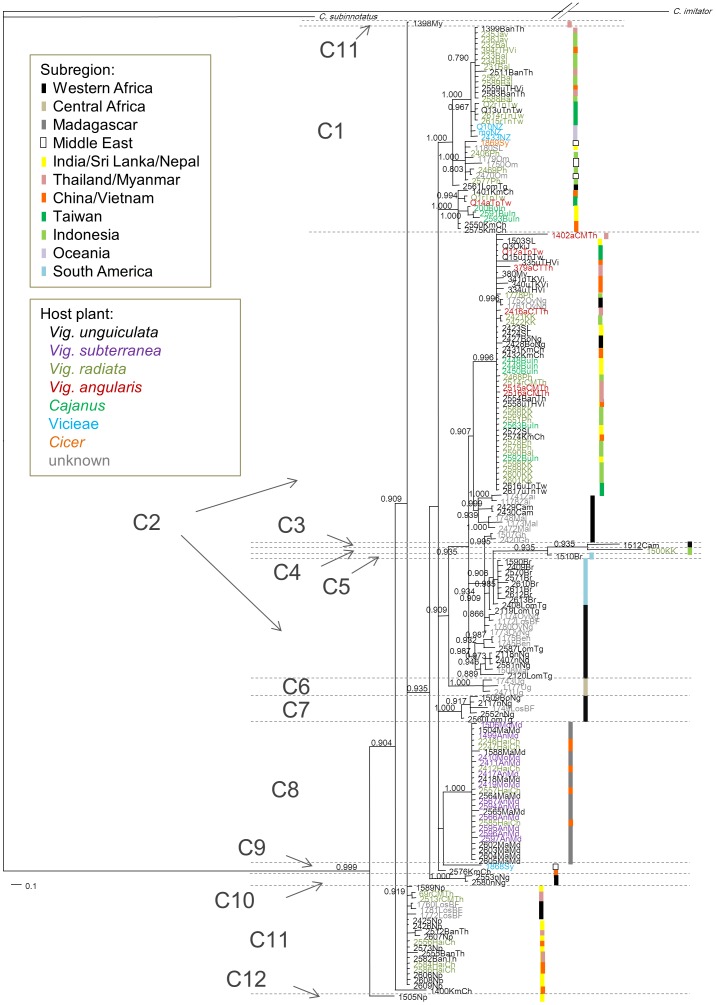
The Bayesian majority rule consensus tree based on mitochondrial genes was polytomized (Fig. 1). Twelve clades (C1–C12), which were diverged with at least 0.5 nucleotide substitutions per site from the closest clade, were recognized. Based on these clades, there have been at least seven introductions of C. maculatus into Asia (clades C1, C2, C4, C8, C9, C11 and C12, Figs. 1 and 2). Separate gene trees for COI and tRNALeu–COII and the combined tree differed in topology but the combined and the COI trees revealed the very same set of clades (Figs. 1 and 1).
Figure 1. Bayesian majority rule consensus tree of Callosobruchus maculatus populations, based on mitochondrial genes.
The numbers near the nodes indicate posterior probabilities (only probabilities >0.700 are shown). The colors of the sample codes indicate the host plants used by the populations when collected. The colors of the vertical bars indicate the geographic regions where the population samples were collected.
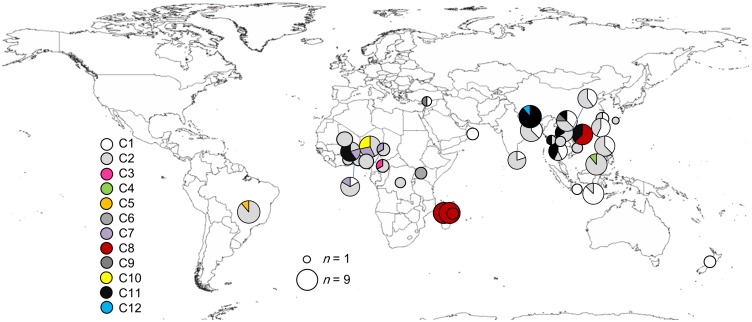
Figure 2. World map showing haplotypic compositions (based on clades identified in Fig. 1) of local populations.
The size of pie charts indicates the number of individuals sampled.
None of the individuals showed a positive diagnostic Wolbachia PCR, in line with the previous finding that Wolbachia infection does not occur in C. maculatus [87].
Congruence between C. maculatus phylogeny and geography/hosts
Randomization of the geographic locations changed the length of the C. maculatus mitochondrial phylogenetic tree (T-PTP, P = 0.001), indicating highly significant congruence between the beetle phylogeny and geographic regions. Inclusion of individuals without host information did not alter the significant relationship between geographic location and the C. maculatus phylogeny (T-PTP, P = 0.001). Randomization of the host bean groups also changed the length of the C. maculatus phylogenetic tree (T-PTP, P = 0.001), indicating that the congruence between the beetle phylogeny and host plant use was as significant as between phylogeny and geography.
Population structure
Out of the 160 individuals sampled recently (Table 1), there were 93 distinct haplotypes for COI (gene diversity ± SD, 0.970±0.007; nucleotide diversity ± SD, 0.0282±0.0137) and, out of 91 sequences, 34 haplotypes for tRNALeu–COII (gene diversity, 0.943±0.010; nucleotide diversity, 0.0165±0.0083). In total, there were 111 different mitochondrial haplotypes (gene diversity, 0.991±0.003; nucleotide diversity, 0.0136±0.0066). For the EF-1α, out of 69 individuals sequenced, there were 60 different alleles (gene diversity ± SD, 0.991±0.006; nucleotide diversity ± SD, 0.0344±0.0169).
The AMOVA on the mitochondrial genes revealed significant genetic differentiation among continents/archipelagos, among subregions within continents/archipelagos, and within subregions among populations (Table 2). However, when unequal sample sizes were corrected by randomly re-sampling two individuals per population, the among-continent/archipelago variation became much less pronounced (P = 0.016–0.072). When continents were controlled for, subregional differentiation was significant in Africa but not in Asia (Table 2). When host plant usage was restricted to only the populations using V. unguiculata, variation across continents was no longer significant (Table 2).
Table 2. Tests of genetic differentiation among and within continents/archipelagos (regions).
| AMOVA (% variance) [d.f.] | |||||
| Global | Continent controlled | Host plants controlled (V. unguiculata) | |||
| mDNA | nDNA | Africa | Asia | ||
| Among continents or archipelagos | 18.3**a [4] | −8.7NS [4] | 8.5 NS [2] | ||
| Among subregions | 15.4** [7] | 24.8** a [6] | 46.3** [2] | −7.4NS [4] | 24.2** a [4] |
| Among populations | 66.3** [148] | 83.9* [57] | 21.3** a [5] | 45.5** a [13] | 67.3** a [71] |
| Within populations | - | - | 32.4** [35] | 61.9** [82] | - |
| Among host plants | 17.0** [4] | 2.5* a [3] | −4.0 NS [1] | 31.6** [2] | - |
Analyses of molecular variance (AMOVAs). Data include populations analyzed immediately after collection or reared in the lab briefly (Table 1). Unless otherwise noted as the nuclear DNA (nDNA), the mitochondrial DNA (mtDNA) sequence data were used.
*P<0.05, **P<0.01, NS: P>0.05.
The statistical significance was not robust to equalizing the sample sizes per population (see text).
The AMOVA indicated genetic differentiation among populations using different host-plant species globally for mitochondrial DNA (Table 2). However, host plant usage and geographic area were associated. Thus, we controlled for continental effects and re-analyzed the data. When geography was thus controlled for, there was no significant genetic differentiation among populations using different host species for African populations (Madagascar). However, significant variation was found among populations using different host plants for Asian populations (Chiang Mai, Taipei, Tainan, and Thanh Hoa, where populations were sampled from multiple host species) (P = 0.0089, Table 2). When all Asian populations were included, the variation across populations remained significant among populations using different host plants (variation 9.8% of total variation, d.f. = 3, P = 0.0029). None of these results changed, in terms of our ability to reject null hypotheses, when unequal sample sizes per population were controlled for.
In contrast to the results based on mitochondrial DNA, the AMOVA of the nuclear gene showed no genetic differentiation among continents. Genetic differentiation among populations using different host plants was significant as in mitochondrial genes but this effect was no longer present when the sample size was controlled for (Table 2).
Isolation by distance
Geographic distance (ln-transformed) and F ST/(1- F ST) between pairwise populations were significantly correlated (Mantel test P = 0.0020, Table 3), supporting the hypothesis that IBD forms at least one component of population differentiation. The correlation was significant both within Africa (P = 0.0305, Table 3) and within Asia (P = 0.0410, Table 3). When host plants were restricted to V. unguiculata, however, the global correlation was marginally non-significant (P = 0.0545, Table 3).
Table 3. Tests of the isolation by distance hypothesis (IBD) for the mitochondrial genes.
| IBD (rS) [n] | ||||
| Global (all regions and host plants included) | Continent controlled | Host plants controlled (V. unguiculata) | ||
| Africa | Asia | |||
| Population | 0.308** [434] | 0.437* [28] | 0.252* [152] | 0.243 NS [105] |
Mantel test P, *:<0.05, **:<0.01, NS: P>0.05. n: number of pairwise comparisons.
Demographic growth and selective neutrality
Since our sample sizes were large, we focused on the FS. We failed to reject a scenario of population stability in the recent past combined with selective neutrality for the mitochondrial genes for global, African or Asian populations and for the populations using either V. unguiculata or other hosts (Table 4). In contrast, for nDNA, selective neutrality was rejected for the global, African, or Asian population and for the population using V. unguiculata (Table 5). The model of a recent past constant population size was rejected for global and African populations for nDNA, indicating demographic growth on the global scale and in Africa in C. maculatus (Table 5). When host plant was controlled to either V. unguiculata or other hosts, recent past global population growth in C. maculatus was no longer supported (Table 5). The number of segregating (polymorphic) sites was higher for African populations than for Asian populations and for populations using V. unguiculata than those using hosts other than V. unguiculata, for nDNA (Table 5).
Table 4. Tests of sudden demographic growth in the recent past for the mitochondrial genes.
| Global | Continent | Host plant | |||
| Africa | Asia | V. unguiculata | Other hosts | ||
| n | 158 | 43 | 99 | 78 | 73 |
| S | 81 | 50 | 88 | 60 | 88 |
| D | −0.511 NS | −0.708 NS | −0.518 NS | −0.236 NS | −0.641 NS |
| Raggedness r | 0.0892 NS | 0.1026 NS | 0.0587NS | 0.0642 NS | 0.0892 NS |
| FS | −0.43 NS | 0.59 NS | 4.82 NS | 0.49 NS | 6.57 NS |
| R 2 | 0.0767 NS | 0.0913NS | 0.0855NS | 0.0985 NS | 0.0858 NS |
Table 5. Tests of sudden demographic growth in the recent past for the nuclear gene.
| Global | Continent | Host plant | |||
| Africa | Asia | V. unguiculata | Other hosts | ||
| n | 68 | 23 | 36 | 28 | 37 |
| S | 11 | 50 | 33 | 82 | 9 |
| D | −1.630* | −1.974** | −2.015** | −2.058** | −1.145 NS |
| Raggedness r | 0.0688 NS | 0.0130* | 0.0198* | 0.0158 NS | 0.0519 NS |
| FS | −6.38** | −5.99* | −0.53 NS | −3.82 NS | −2.39 NS |
| R 2 | 0.0461* | 0.0556*** | 0.0966 NS | 0.0570** | 0.0744 NS |
Discussion
We aimed to study the relative contribution of factors associated with geography, host plant usage and human-aided dispersal to genetic population structure of the stored bean pest, C. maculatus, mainly in Africa and Asia. On a continental scale, there was among-subregional genetic variation within Africa but not within Asia. Among-host genetic population differences were found only in Asian C. maculatus populations. The IBD hypothesis was supported globally, but not for the populations using V. unguiculata, the main host of C. maculatus. The number of segregating sites was higher for populations in Africa than in Asia and for those using hosts other than V. unguiculata than those using V. unguiculata for the nuclear gene, supporting the hypothesis that Africa is the region of origin for C. maculatus [45]. We suggest that the differentiation among hosts in C. maculatus is due to multiple invasions on novel hosts by different alleles, particularly in Asia. Geographic effects [8] that are larger than host-plant effects [10] have also been reported in cosmopolitan moths, the apple moth and the fruit moth, although among-continental differentiation seems more pronounced in those pests than in C. maculatus (Table 2). This may be partly due to a higher frequency of anthropogenic cross-continental dispersal in C. maculatus, aided by human trade of host beans than in these moths. Different levels of dispersal may be generated by differences in the frequency of inter-continental trade, in pest control efficiency and in pest survival during transport (fresh, soft, decaying fruits in moths versus dry, hard seeds in C. maculatus).
Geographic structuring
We found that there was genetic structure among African C. maculatus subregional populations and suggest that this most likely represents allopatric divergence, because the structure primarily reflects the geographic separation of populations (Figs. 1 and 2, Table 2). This is likely mainly the result of historical differentiation along with natural and human-aided dispersal [88] and more recent international trade of dry cowpeas, V. unguiculata, which has been restricted mostly to within Western Africa [49]. We note that it is also possible that some degree of local adaptation and/or reproductive incompatibility between different geographic populations of C. maculatus has contributed to the maintenance of geographic genetic structure. Asian populations show limited mating incompatibility (with African populations [89],[90]) as well as some prezygotic postmating incompatibility (with a Brazilian population [91]).
We could not reject the hypothesis that the mtDNA variation documented here is primarily due to genetic drift (Table 4). This may seem surprising given that phenotypic effects of mtDNA variation have been demonstrated in C. maculatus [40],[92] and in many other species [93],[94]. However, geographically varying natural selection regimes, caused for example by differences in thermal environments, could contribute to patterns such as that documented here. Moreover, the fact that within-population mtDNA haplotype variation was found to be fairly high and significant (Table 2) is interesting in light of recent experimental work in C. maculatus demonstrating that negative frequency-dependent selection acts to maintain within-population genetic variation of mtDNA haplotypes in the laboratory [95]. Although our data do not allow for formal tests of frequency dependent selection, this finding suggests that natural selection may play a role in differentiation of mitochondrial genetic variation.
Host plant race
Host race formation likely contributes to the maintenance of genetic structure in invaded areas like Asia (Table 2). However, host acceptance of C. maculatus is broad and evolves rapidly in the laboratory (e.g., [34],[96],[97]) and this broad and labile host acceptance may impede the formation of rigid host-races. The high genetic diversity among populations using different hosts in Asia is thus intriguing. One might also argue that selection and cultivation of Asian yardlong bean, V. unguiculata var. sesquipedalis could have contributed to the genetic differentiation of this seed-predator in Asia. However, seedpods of yardlong beans are harvested at a young stage to use as a fresh vegetable [49], unlike other V. unguiculata varieties that are harvested at later mature stages for dry seeds, and this most likely limits infestation by postharvest pests such as C. maculatus. We suggest that the differentiation among populations using different host plants in Asia may be the combined result of multiple invasions by different alleles followed by host-shifting to Asian legumes, although it is difficult to exclude a role of selection. The reduced cultivation of V. unguiculata relative to other beans in Asia likely decreased the availability of V. unguiculata for C. maculatus and may increase the chance of exploration and further differentiation on alternative hosts.
Anthropogenic dispersal
Overall, genetic differentiation in C. maculatus was primarily due to geographic isolation and multiple cross-continental anthropogenic dispersal events rather than to the formation of strict host-plant races. More frequent within-continental international trade of dry beans likely reduced the subregional genetic differentiation in Asia more than in Africa. In spite of the overall geographic structuring, some Asian populations shared similar haplotypes with African populations (e.g., Hainan and Madagascar populations, clade C8; India, Sri Lanka, Myanmar, Thailand, China, Japan, Taiwan, Philippines and Nigeria, clade C2 in Figs. 1 and 2). This may represent examples of more recent introductions into Asia from Africa. Three million out of 3.6 million tonnes of annual world production of dry cowpeas were produced in Western Africa during 1999–2003, the main producer being Nigeria [47],[49],[98]. Since the mid-1990s, Africa has exported/imported negligible amounts of dried cowpeas (Fig. S2, [97]). Furthermore, about 80% of the world cowpea trade takes place within Western and Central Africa [99]. Likewise, international trade of dry cowpeas and other Vigna beans within Asia probably masked any effect of geographic isolation (e.g., between ASEAN countries [Association of Southeast Asian Nations], the main exporter being Myanmar, [97],[100]).
To conclude, we found significant subregional-scale genetic differentiation in Africa and among-host difference in Asia. We suggest that this contrasting pattern has primarily been caused by allopatric differentiation in Africa and by multiple invasions by different alleles, followed by host shifts in Asia. Recent long-range human trade of dry beans has allowed dispersal from Africa to other continents and medium-range trade has subsequently intermixed the introduced alleles within Asia. In Africa, intermixing has occurred at a smaller spatial scale.
Supporting Information
Bayesian majority rule consensus tree of Callosobruchus maculatus populations, based only on the mitochondrial COI gene. The numbers near the nodes indicate posterior probabilities (only probabilities >0.700 are shown).
(TIF)
Export and import quantities of dry cowpeas (tonnes) in different African and Asian subregions. Data on imports into South Asia were unavailable.
(TIF)
Acknowledgments
We thank J. Adorada, T.C. Banerjee, S. Buranapanichpan, P. Credland, T.D. Hoa, G. Adolé, H. Ishikawa, S.M. Mehrez, B. Napompeth, T. Ofuya, K. Ogata, M. Shimada and T. Yamanaka for providing specimens.
Funding Statement
This study was supported by Grants-in-Aid from JSPS (http://www.jsps.go.jp/english/index.html) (17405005, 20405006, 19510237, 22570215, 25430194 and 23405008) to MT and YT and by grants from the Swedish Research Council (http://www.vr.se/inenglish.4.12fff4451215cbd83e4800015152.html) (621-2010-5266) and the European Research Council (http://erc.europa.eu/) (AdG-294333) to GA. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Downie DA, Fisher JR, Granett J (2001) Grapes, galls, and geography: The distribution of nuclear and mitochondrial DNA variation across host-plant species and regions in a specialist herbivore. Evolution 55: 1345–1362. [PubMed] [Google Scholar]
- 2. Ruiz EA, Rinehart JE, Hayes JL, Zuniga G (2009) Effect of geographic isolation on genetic differentiation in Dendroctonus pseudotsugae (Coleoptera: Curculionidae). Hereditas 146: 79–92. [DOI] [PubMed] [Google Scholar]
- 3. Kerdelhue C, Roux-Morabito G, Forichon J, Chambon JM, Robert E, et al. (2002) Population genetic structure of Tomicus piniperda L. (Curculionidae: Scolytinae) on different pine species and validation of T. destruens (Woll.). Mol Ecol 11: 483–494. [DOI] [PubMed] [Google Scholar]
- 4. Scheffer SJ, Hawthorne DJ (2007) Molecular evidence of host-associated genetic divergence in the holly leafminer Phytomyza glabricola (Diptera: Agromyzidae): apparent discordance among marker systems. Mol Ecol 16: 2627–2637. [DOI] [PubMed] [Google Scholar]
- 5. Behere GT, Tay WT, Russell DA, Heckel DG, Appleton BR, et al. (2007) Mitochondrial DNA analysis of field populations of Helicoverpa armigera (Lepidoptera: Noctuidae) and of its relationship to H. zea . BMC Evol Biol 7: 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tuda M, Wasano N, Kondo N, Horng S-B, Chou L-Y, et al. (2004) Habitat-related mtDNA polymorphism in the stored-bean pest Callosobruchus chinensis (Coleoptera: Bruchidae). Bull Entomol Res 94: 75–80. [DOI] [PubMed] [Google Scholar]
- 7. Thaler R, Brandstatter A, Meraner A, Chabicovski M, Parson W, et al. (2008) Molecular phylogeny and population structure of the codling moth (Cydia pomonella) in Central Europe: II. AFLP analysis reflects human-aided local adaptation of a global pest species. Mol Phylogenet Evol 48: 838–849. [DOI] [PubMed] [Google Scholar]
- 8. Tooman LK, Rose CJ, Carraher C, Suckling DM, Paquette SR, et al. (2011) Patterns of mitochondrial haplotype diversity in the invasive pest Epiphyas postvittana (Lepidoptera: Tortricidae). J Econ Entomol 104: 920–932. [DOI] [PubMed] [Google Scholar]
- 9. Margaritopoulos JT, Kasprowicz L, Malloch GL, Fenton B (2009) Tracking the global dispersal of a cosmopolitan insect pest, the peach potato aphid. BMC Ecology 9: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kirk H, Dorn S, Mazzi D (2013) Worldwide population genetic structure of the oriental fruit moth (Grapholita molesta), a globally invasive pest. BMC Ecology 13: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Southgate BJ (1981) Univoltine and multivoltine cycles: their significance. In: Labeyrie V, editor. The ecology of bruchids attacking legumes (pulses). The Hague: Dr. W. Junk Publishers. pp. 17–22. [Google Scholar]
- 12. Ishihara M (1998) Geographical variation in insect developmental period: effect of host plant phenology on the life cycle of the bruchid seed feeder Kytorhinus sharpianus . Entomol Exp Appl 87: 311–319. [Google Scholar]
- 13. Alvarez N, McKey D, Hossaert-McKey M, Born C, Mercier L, et al. (2005) Ancient and recent evolutionary history of the bruchid beetle, Acanthoscelides obtectus Say, a cosmopolitan pest of beans. Mol Ecol 14: 1015–1024. [DOI] [PubMed] [Google Scholar]
- 14. Tuda M, Ronn J, Buranapanichpan S, Wasano N, Arnqvist G (2006) Evolutionary diversification of the bean beetle genus Callosobruchus (Coleoptera: Bruchidae): traits associated with stored-product pest status. Mol Ecol 15: 3541–3551. [DOI] [PubMed] [Google Scholar]
- 15. Kato T, Bonet A, Yoshitake H, Romero-Napoles J, Jinbo U, et al. (2010) Evolution of host utilization patterns in the seed beetle genus Mimosestes Bridwell (Coleoptera: Chrysomelidae: Bruchinae). Mol Phylogenet Evol 55: 816–832. [DOI] [PubMed] [Google Scholar]
- 16. Tuda M, Chou L-Y, Niyomdham C, Buranapanichpan S, Tateishi Y (2005) Ecological factors associated with pest status in Callosobruchus (Coleoptera: Bruchidae): high host specificity of non-pests to Cajaninae (Fabaceae). J Stored Prod Res 41: 31–45. [Google Scholar]
- 17. Benard MF, McCauley SJ (2008) Integrating across life-history stages: consequences of natal habitat effects on dispersal. Am Nat 171: 553–567. [DOI] [PubMed] [Google Scholar]
- 18.Shinoda K, Yoshida T (1990) The life history of the azuki bean weevil, Callosobruchus chinensis L. (Coleoptera: Bruchidae), in the field. In: Fujii K, Gatehouse AMR, Johnson CD, Mitchel R, Yoshida T, editors. Bruchids and legumes: economics, ecology and coevolution. Dordrecht: Kluwer. pp. 149–159. [Google Scholar]
- 19. Yoshimoto CM, Gressitt JL (1959) Trapping of air-borne insects on ships on the Pacific, Part 2. Proc Hawaii Entomol Soc 17: 150–155. [Google Scholar]
- 20. Harrell JC, Holzapfel E (1966) Trapping of air-borne insects on ships in the Pacific, Part 6. Pac Insects 8: 33–42. [Google Scholar]
- 21. Holzapfel EP, Perkins BD Jr (1969) Trapping of air-borne insects on ships in the Pacific, Part 7. Pac Insects 11: 455–476. [Google Scholar]
- 22. Ouedraogo PA, Monge JP, Huignard J (1991) Importance of temperature and seed water content on the induction of imaginal polymorphism in Callosobruchus maculatus . Entomol Exp Appl 59: 59–66. [Google Scholar]
- 23. Nahdy MS, Silim SN, Ellis RH (1999) Effect of field infestations of immature pigeonpea (Cajanus cajan (L.) Millsp.) pods on production of active (flight) and sedentary (flightless) morphs of Callosobruchus chinensis (L.). J Stored Prod Res 35: 339–354. [Google Scholar]
- 24. Caswell GH (1986) The value of the pod in protecting cowpea seed from attack by bruchid beetles. Samaru J Agr Res 2: 49–55. [Google Scholar]
- 25.Murdock LL, Shade RE, Kitch LW, Ntoukam G, Lowenberg-DeBoer J, et al.. (1997) Postharvest storage of cowpea in sub-Saharan Africa. In: Singh BB, Mohan Raj DR, Dashiell KE, Jackai LEN, editors. Advances in cowpea research. Ibadan and Ibaraki: IITA and JIRCAS. pp. 302–312. [Google Scholar]
- 26. Southgate BJ, Howe R, Brett GA (1957) The specific status of Callosobruchus maculatus (F.) and Callosobruchus analis (F.). Bull Entomol Res 48: 78–89. [Google Scholar]
- 27. Utida S (1972) Density dependent polymorphism in adult of Callosobruchus maculatus (Coleoptera, Bruchidae). J Stored Prod Res 8: 111–126. [Google Scholar]
- 28. Janzen DH (1977) How southern cowpea weevil larvae (Bruchidae: Callosobruchus maculatus) die on non-host seeds. Ecology 58: 921–927. [Google Scholar]
- 29. Fujii K (1968) Studies on interspecies competition between the azuki bean weevil and the southern cowpea weevil, 3. some characteristics of strains of two species. Res Popul Ecol 10: 87–98. [Google Scholar]
- 30. Bellows TS, Hassell MP (1984) Models for interspecific competition in laboratory populations of Callosobruchus spp. J Anim Ecol 53: 831–848. [Google Scholar]
- 31.Fujii K, Gatehouse AMR, Johnson CD, Mitchel R, Yoshida T (1990) Bruchids and legumes: economics, ecology and coevolution. Dordrecht: Kluwer. 407 p. [Google Scholar]
- 32.Mitchell R (1990) Behavioral ecology of Callosobruchus maculatus. In: Fujii K, Gatehouse AMR, Johnson CD, Mitchel R, Yoshida T, editors. Bruchids and legumes: economics, ecology and coevolution. Dordrecht: Kluwer. pp. 317–330. [Google Scholar]
- 33. Toquenaga Y (1993) Contest and scramble competitions in Callosobruchus maculatus (Coleoptera: Bruchidae), 2. larval competition and interference mechanisms. Res Popul Ecol 35: 57–68. [Google Scholar]
- 34. Messina FJ (2004) How labile are the egg-laying preferences of seed beetles? Ecol Entomol 29: 318–326. [Google Scholar]
- 35. Stillwell RC, Wallin WG, Hitchcock LJ, Fox CW (2007) Phenotypic plasticity in a complex world: interactive effects of food and temperature on fitness components of a seed. Oecologia 153: 309–321. [DOI] [PubMed] [Google Scholar]
- 36. Nakamichi Y, Toquenaga Y, Fujii K (2008) Persistent host-parasitoid interaction caused by host maturation variability. Popul Ecol 50: 191–196. [Google Scholar]
- 37. Tuda M (1996) Temporal/spatial structure and the dynamical property of laboratory host-parasitoid systems. Res Popul Ecol 38: 133–140. [Google Scholar]
- 38. Tuda M (2011) Evolutionary diversification of bruchine beetles: climate-dependent traits and development associated with pest status. Bull Entomol Res 101: 415–422. [DOI] [PubMed] [Google Scholar]
- 39. Angus RB, Dellow J, Winder C, Credland PF (2011) Karyotype differences among four species of Callosobruchus Pic (Coleoptera: Bruchidae). J Stored Prod Res 47: 76–81. [Google Scholar]
- 40. Arnqvist G, Dowling DK, Eady P, Gay L, Tregenza T, et al. (2010) The genetic architecture of metabolic rate: environment specific epistasis between mitochondrial and nuclear genes in an insect. Evolution 64: 3354–3363. [DOI] [PubMed] [Google Scholar]
- 41. Mano H, Toquenaga Y (2011) Contest-type competition between age classes in scramble-type Callosobruchus maculatus (Coleoptera: Bruchidae). Entomol Sci 14: 166–172. [Google Scholar]
- 42. Vamosi SM, den Hollander MD, Tuda M (2011) Egg dispersion is more important than competition type for herbivores attacked by a parasitoid. Popul Ecol 53: 319–326. [Google Scholar]
- 43. Ishii Y, Shimada M (2012) Learning predator promotes coexistence of prey species in host-parasitoid systems. Proc Nat Acad Sci USA 109: 5116–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Messina FJ, Pena NM (2012) Mode of inheritance of increased host acceptance in a seed beetle. Bull Entomol Res 102: 497–503. [DOI] [PubMed] [Google Scholar]
- 45. Anton K-W, Halperin J, Calderon M (1997) An annotated list of the Bruchidae (Coleoptera) of Israel and adjacent areas. Isr J Entomol 31: 59–96. [Google Scholar]
- 46. Mbar Tine E, Diome T, Kebe K, Ndong A, Douma A, et al. (2013) Genetic diversity of Callosobruchus maculatus Fabricus (cowpea weevil) populations in various agro-ecological areas of five countries in West African sub-region. South Asian J Exp Biol 3: 71–83. [Google Scholar]
- 47.Quin FM (1997) Introduction. In: Singh BB, Mohan Raj DR, Dashiell KE, Jackai LEN, editors. Advances in Cowpea Research. Ibadan and Ibaraki: IITA and JIRCAS. pp. ix–xv. [Google Scholar]
- 48. Ehlers JD, Hall AE (1997) Cowpea (Vigna unguiculata L. Walp.). Field Crops Res 53: 187–204. [Google Scholar]
- 49.Madamba R, Grubben GJH, Asante IK, Akromah R (2006) Vigna unguiculata (L.) Walp. In: Brink M, Belay G, editors. Plant resources of tropical Africa 1. cereals and pulses. Wageningen: PROTA Foundations. pp. 221–229. [Google Scholar]
- 50. Coulibaly S, Pasquet RS, Papa R, Gepts P (2002) AFLP analysis of the phenetic organization and genetic diversity of Vigna unguiculata L. Walp. reveals extensive gene flow between wild and domesticated types. Theor Appl Genet 104: 358–366. [DOI] [PubMed] [Google Scholar]
- 51. Ba FS, Pasquet RS, Gepts P (2004) Genetic diversity in cowpea [Vigna unguiculata (L.) Walp.] as revealed by RAPD markers. Genet Resour Crop Evol 51: 539–550. [Google Scholar]
- 52.Ng NQ, Maréchal R (1985) Cowpea taxonomy, origin and germplasm. In: Singh S, Rachie K, editors. Cowpea research, production, and utilization. Chichester: Wiley. pp. 11–21. [Google Scholar]
- 53. Tomooka N, Kashiwaba K, Vaughan DA, Ishimoto M, Egawa Y (2000) The effectiveness of evaluating wild species: searching for sources of resistance to bruchid beetles in the genus Vigna subgenus Ceratotropis . Euphytica 115: 27–41. [Google Scholar]
- 54. Arora GL (1977) Bruchidae of northwest India. Oriental Insects Suppl. 7 1–132. [Google Scholar]
- 55. Desroches P, Elshazly E, Mandon N, Duc G, Huignard J (1995) Development of Callosobruchus chinensis (L) and C. maculatus (F) (Coleoptera, Bruchidae) in seeds of Vicia faba L differing in their tannin, vicine and convicine contents. J Stored Prod Res 31: 83–89. [Google Scholar]
- 56. Gomez-Zurita J, Juan C, Petitpierre E (2000) The evolutionary history of the genus Timarcha (Coleoptera, Chrysomelidae) inferred from mitochondrial COII gene and partial 16S rDNA sequences. Mol Phylogenet Evol 14: 304–317. [DOI] [PubMed] [Google Scholar]
- 57. Simon C, Frati F, Beckenbach A, Crespi B, Liu H, et al. (1994) Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved polymerase chain reaction primers. Ann Entomol Soc Am 87: 651–701. [Google Scholar]
- 58. Mardulyn P, Othmezouri N, Mikhailov YE, Pasteels JM (2011) Conflicting mitochondrial and nuclear phylogeographic signals and evolution of host-plant shifts in the boreo-montane leaf beetle Chrysomela lapponica . Mol Phylogenet Evol 61: 686–696. [DOI] [PubMed] [Google Scholar]
- 59. Kim C-G, Zhou H-Z, Imura Y, Tominaga O, Su Z-H, et al. (2000) Pattern of morphological diversification in the Leptocarabus ground beetles (Coleoptera: Carabidae) as deduced from mitochondrial ND5 gene and nuclear 28S rDNA sequences. Mol Biol Evol 17: 137–145. [DOI] [PubMed] [Google Scholar]
- 60. Belshaw R, Quicke DLJ (2002) Robustness of ancestral state estimates: evolution of life history strategy in ichneumonoid parasitoids. Syst Biol 51: 450–477. [DOI] [PubMed] [Google Scholar]
- 61. Turelli M, Hoffmann AA (1991) Rapid spread of an inherited incompatibility factor in California Drosophila . Nature 353: 440–442. [DOI] [PubMed] [Google Scholar]
- 62. Holden PR, Brookfield JFY, Jones P (1993) Cloning and characterization of an ftsZ homologue from a bacterial symbiont of Drosophila melanogaster . Mol Gen Genet 240: 213–220. [DOI] [PubMed] [Google Scholar]
- 63. Kondo N, Shimada M, Fukatsu T (1999) High prevalence of Wolbachia in the azuki bean beetle Callosobruchus chinensis (Coleoptera, Bruchidae). Zool Sci 16: 955–962. [Google Scholar]
- 64. Huelsenbeck JP, Ronquist F (2001) MrBayes: Bayesian inference of phylogeny. Biometrics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- 65. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinfomatics 19: 1572–1573. [DOI] [PubMed] [Google Scholar]
- 66.Nylander JAA (2004) MrAIC.pl. Program distributed by the author. Uppsala: Evolutionary Biology Centre, Uppsala Univ. [Google Scholar]
- 67. Hasegawa M, Kishino H, Yano T (1985) Dating of human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol 22: 160–174. [DOI] [PubMed] [Google Scholar]
- 68.Geyer CJ (1991) Markov chain Monte Carlo maximum likelihood. In: Keramidas EM, editor. Computing science and statistics. Proceedings of the 23rd symposium on the interface. Seattle: Interface Foundation of North America. pp. 156–163. [Google Scholar]
- 69. Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E (1953) Equation of state calculations by fast computing machines. J Chem Phys 21: 1087–1092. [Google Scholar]
- 70. Hastings WK (1970) Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57: 97–109. [Google Scholar]
- 71.Rambaut A, Drummond AJ (2009) Tracer v1.5. Available: http://beast.bio.ed.ac.uk/. [Google Scholar]
- 72.Swofford DL (2002) PAUP*. Phylogenetic analysis using parsimony (and other methods), 4.0 beta. Sunderland: Sinauer Associates Inc. [Google Scholar]
- 73. Trueman JWH (1996) Permutation tests and outgroups. Cladistics 12: 253–261. [DOI] [PubMed] [Google Scholar]
- 74. Cockerham CC (1969) Variance of gene frequencies. Evolution 23: 72–83. [DOI] [PubMed] [Google Scholar]
- 75. Cockerham CC (1973) Analysis of gene frequencies. Genetics 74: 679–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weir BS, Cockerham CC (1984) Estimating F-statistics population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- 77. Excoffier L, Lischer H (2010) Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10: 564–567. [DOI] [PubMed] [Google Scholar]
- 78. Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10: 512–526. [DOI] [PubMed] [Google Scholar]
- 79. Wright S (1943) Isolation by distance. Genetics 28: 114–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86: 248–249. [Google Scholar]
- 81. Rousset F (2008) Genepop'007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8: 103–106. [DOI] [PubMed] [Google Scholar]
- 82. Harpending H (1994) Signature of ancient population growth in a low-resolution mitochondrial DNA mismatch distribution. Human Biol 66: 591–600. [PubMed] [Google Scholar]
- 83. Fu Y-X (1997) Statistical tests of neutrality of mutations against population growth, hitchhiking and background selection. Genetics 147: 915–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Ramos-Onsins SE, Rozas J (2002) Statistical properties of new neutrality tests against population growth. Mol Biol Evol 19: 2092–2100. [DOI] [PubMed] [Google Scholar]
- 85. Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25: 1451–1452. [DOI] [PubMed] [Google Scholar]
- 86. Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123: 585–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Kondo NI, Tuda M, Toquenaga Y, Lan Y-C, Buranapanichpan S, et al. (2011) Wolbachia infections in world populations of bean beetles (Coleoptera: Chrysomelidae: Bruchinae) infesting cultivated and wild legumes. Zool Sci 28: 501–508. [DOI] [PubMed] [Google Scholar]
- 88. Diamond J (1997) Guns, germs, and steel: the fates of human societies. Norton & Company, New York [Google Scholar]
- 89. Brown DV, Eady PE (2001) Functional incompatibility between the fertilization systems of two allopatric populations of Callosobruchus maculatus (Coleoptera: Bruchidae). Evolution 55: 2257–2262. [DOI] [PubMed] [Google Scholar]
- 90. Messina FJ, Bloxham AJ, Seargent AJ (2007) Mating compatibility between geographic populations of the seed beetle Callosobruchus maculatus . J Insect Behav 20: 489–501. [Google Scholar]
- 91. Fricke C, Arnqvist G (2004) Divergence in replicated phylogenies: the evolution of partial post-mating prezygotic isolation in bean weevils. J Evol Biol 17: 1345–1354. [DOI] [PubMed] [Google Scholar]
- 92. Dowling DK, Abiega KC, Arnqvist G (2007) Temperature-specific outcomes of cytoplasmic-nuclear interactions on egg-to-adult development time in seed beetles. Evolution 61: 194–201. [DOI] [PubMed] [Google Scholar]
- 93. Ehinger M, Fontanillas P, Petit E, Perrin N (2002) Mitochondrial DNA variation along an altitudinal gradient in the greater white-toothed shrew, Crocidura russula . Mol Ecol 11: 939–945. [DOI] [PubMed] [Google Scholar]
- 94. Dowling DK, Friberg U, Lindell J (2008) Evolutionary implications of non-neutral mitochondrial genetic variation. Trends Ecol Evol 23: 546–554. [DOI] [PubMed] [Google Scholar]
- 95. Kazancıoğlu E, Arnqvist G (2014) The maintenance of mitochondrial genetic variation by negative frequency-dependent selection. Ecol Lett 17: 22–27. [DOI] [PubMed] [Google Scholar]
- 96. Fricke C, Arnqvist G (2007) Rapid adaptation to a novel host in a seed beetle (Callosobruchus maculatus): the role of sexual selection. Evolution 61: 440–454. [DOI] [PubMed] [Google Scholar]
- 97. Messina FJ, Jones JC, Mendenhall M, Muller A (2009) Genetic modification of host acceptance by a seed beetle, Callosobruchus maculatus (Coleoptera: Bruchidae). Ann Entomol Soc Am 102: 181–188. [Google Scholar]
- 98.FAO Statistics Division (2012) FAOSTAT. http://faostat.fao.org, Food and Agriculture Organization of the United Nations.
- 99. Langyintuo AS, Lowenberg-DeBoer J, Faye M, Lambert D, Ibro G, et al. (2003) Cowpea supply and demand in west and central Africa. Field Crops Res 82: 215–231. [Google Scholar]
- 100. United States International Trade Commission (2010) ASEAN: regional trends in economic integration, export competitiveness, and inbound investment for selected industries. Investigation no. 332–511, USITC Publication 4176, 222 pp [Google Scholar]
- 101. Devereau AD, Gudrups I, Appleby JH, Credland PF (2003) Automatic, rapid screening of seed resistance in cowpea, Vigna unguiculata (L.) Walpers, to the seed beetle Callosobruchus maculatus (F.) (Coleoptera: Bruchidae) using acoustic monitoring. J Stored Prod Res 39: 117–129. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Bayesian majority rule consensus tree of Callosobruchus maculatus populations, based only on the mitochondrial COI gene. The numbers near the nodes indicate posterior probabilities (only probabilities >0.700 are shown).
(TIF)
Export and import quantities of dry cowpeas (tonnes) in different African and Asian subregions. Data on imports into South Asia were unavailable.
(TIF)