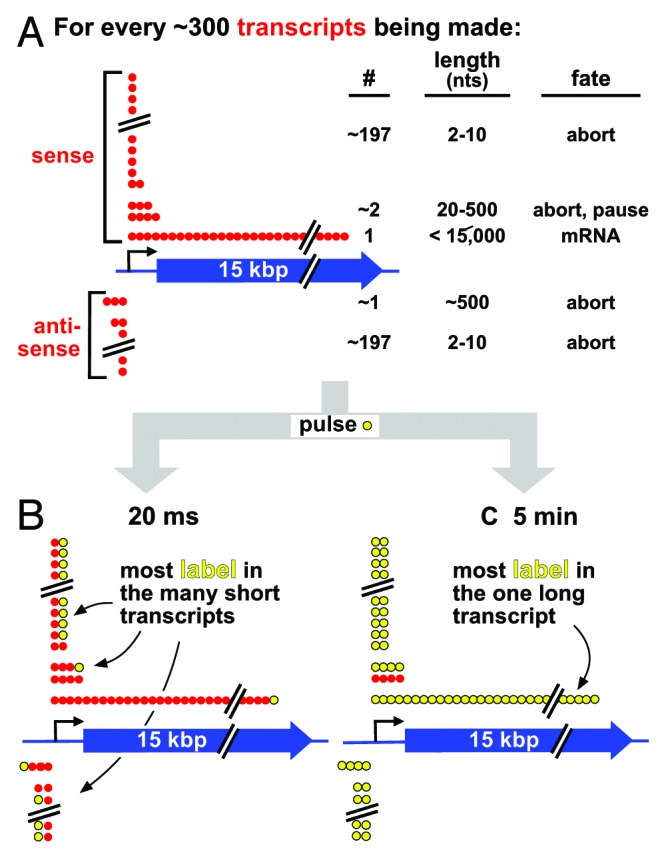
Figure 1. The inefficiency of transcription, and a thought experiment. (A) Strings of red spheres depict nucleotides (nts) incorporated into ~300 transcripts being copied from sense/anti-sense strands of a 15-kbp gene in a cell population. Most polymerases abort prematurely to yield short unstable transcripts of 2–10 nucleotides from both strands, a few pause/abort after generating unstable transcripts of 20–500 nucleotides, and only one generates a stable full-length transcript. The numbers of transcripts in each class are not known, and are included for illustrative purposes only; however, there are probably so many short aborting ones of 2–10 nucleotides that the same general conclusion can be drawn irrespective of precise numbers. (B) After 20 ms (when an elongating polymerase incorporates ~1 labeled nucleotide; yellow), most incorporated label is in short transcripts that soon abort and are quickly degraded; the long transcript contains one label, which goes undetected using most current methods. (C) After 5 min (when a polymerase incorporates ~15 000 nucleotides), the one stable transcript is completely filled with label; however, label incorporated early during the pulse into the many short aborting transcripts has turned over (and so goes undetected).
