Abstract
Small regulatory RNAs (sRNAs) that act by base-pairing were first discovered in so-called accessory DNA elements—plasmids, phages, and transposons—where they control replication, maintenance, and transposition. Since 2001, a huge body of work has been performed to predict and identify sRNAs in a multitude of bacterial genomes. The majority of chromosome-encoded sRNAs have been investigated in E. coli and other Gram-negative bacteria. However, during the past five years an increasing number of sRNAs were found in Gram-positive bacteria. Here, we outline our current knowledge on chromosome-encoded sRNAs from low-GC Gram-positive species that act by base-pairing, i.e., an antisense mechanism. We will focus on sRNAs with known targets and defined regulatory mechanisms with special emphasis on Bacillus subtilis.
Keywords: small regulatory RNA, base-pairing sRNA, low GC Gram-positive bacteria, Bacillus subtilis, Streptococcus pneumoniae
Introduction
Bacterial small regulatory RNAs (sRNAs) are the most abundant class of post-transcriptional regulators and had first been discovered in plasmids, phages, and transposons, where they control replication, maintenance, and transposition.1 Whereas until 2001, only a dozen riboregulators were known from bacterial chromosomes, since then, various systematic approaches have been performed to predict and identify chromosome-encoded sRNAs in prokaryotes. The majority of them have been investigated in E. coli and Salmonella, whereas only a few well-studied examples are known from Gram-positive bacteria.
In the past five years, a combination of computational predictions, transcriptome analyses, and RNA sequencing approaches has been applied to identify chromosome-encoded riboregulators in low GC Gram-positive bacteria (see Table 1). Five searches have been performed in B. subtilis. Rasmussen et al. found 84 putative non-coding trans-encoded sRNAs in the B. subtilis genome,2 and Irnov et al. increased the total number to 108.3 A recent RNA-Seq in B. licheniformis identified 461 independently transcribed sRNAs in addition to 855 RNAs transcribed in antisense to known protein and RNA encoding genes.4 Six searches were performed in S. aureus and five in Listeria monocytogenes, three in Streptococcus pyogenes, and four in Streptococcus pneumoniae (see below). Two searches in Enterococcus faecalis identified 29 novel sRNAs, among them an antisense RNA to 6S RNA.5,6 In Clostridium, three searches have been performed (C. difficile,7 C. ljungdahlii,8 and C. acetobutylicum9).
Table 1. Systematic searches for sRNAs in low GC Gram-positive bacteria.
| Species | sRNAs predicted/confirmed* | References |
|---|---|---|
| Bacillus subtilis | 108 | 2, 3, 97-99 |
| Bacillus licheniformis | 461 indepently encoded | 4 |
| Staphyloccus aureus | 100 trans-,100 cis-encoded | 50, 54, 99, 100-103 |
|
Listeria monocytogenes/ Listeria innocua |
113 trans-, 70 cis-encoded | 72, 104-107 |
| Enterococcus faecalis | 69 | 5, 6 |
| Clostridium difficile | 94 trans-, 91 cis-encoded | 7 |
| Clostridium acetobutylicum | 159 | 9 |
| Clostridium ljungdahlii | 36 | 8 |
| Streptococcus pyogenes | 75 | 84, 108, 109 |
| Streptococcus pneumoniae | 179 | 78, 80, 82, 110 |
The total number of predicted or confirmed sRNAs for the corresponding organisms from all published searches is indicated.
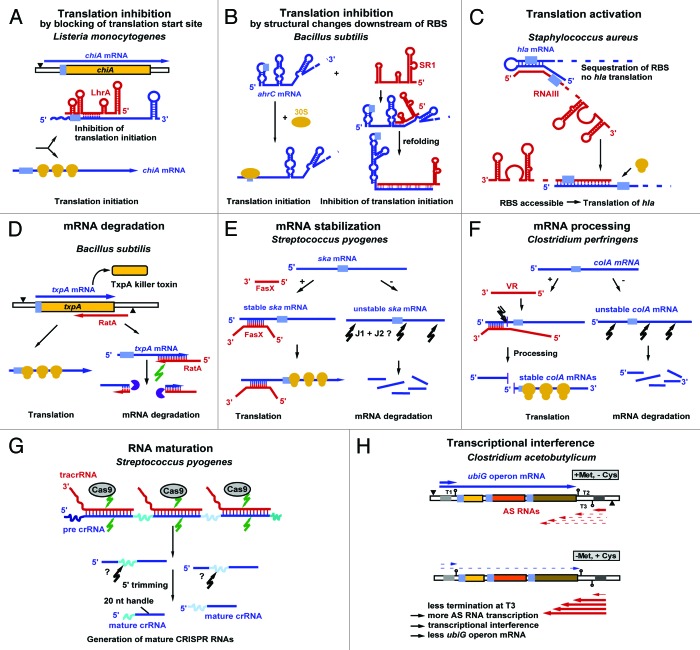
sRNAs either regulate translation or RNA stability. The majority of them inhibit translation (Fig. 1A and B), whereas only a few of them activate translation (Fig. 1C). Translational inhibition can principally occur in three different ways, (1) by direct blocking of the ribosome-binding site (RBS) (Fig. 1A), by induction of structural alterations downstream of the RBS (see Fig. 1B or iii) by blocking of a ribosome standby site required for efficient translation (reviewed in ref. 10). So far, the latter case has been only found in E. coli.11 In some cases, translational inhibition is accompanied by mRNA degradation (see below, S. aureus sRNAs). Both cis-and trans-encoded sRNAs can inhibit translation (Fig. 1A) or promote RNA degradation (Fig. 1D). Some sRNAs can stabilize their target RNAs (Fig. 1E), whereas others process an unstable mRNA into a stable, translationally active RNA (Fig. 1F). In 2011, the first sRNA required for the maturation of long RNAs—the CRISPR RNAs—was discovered12 (Fig. 1G). A mechanism of action that can be exclusively used by cis-encoded sRNAs is transcriptional interference (Fig. 1H, see below). In this report, we summarize (Fig. 1) and discuss all currently known mechanisms employed by chromosome-encoded sRNAs from low-GC Gram-positive bacteria. Thereby, mechanisms discovered for cis-encoded and for trans-encoded sRNAs were assembled. Tables 2 and 3 list all currently known sRNAs with their targets, mechanisms of action, and regulators. In a recent report that also includes plasmid-encoded antisense RNAs that control replication or maintenance of these accessory DNA elements, two additional control mechanisms were considered: transcription and translation attenuation.13 Although both mechanisms are employed by riboswitches, they have not yet been observed for chromosome-encoded bona fide sRNAs.
Figure 1. Mechanisms employed by sRNAs encoded from low-GC Gram-positive bacteria. All currently known mechanisms for sRNAs encoded from chromosomes are summarized. For additional mechanisms employed by plasmid-encoded sRNAs, see reference 13. Antisense RNAs are drawn in red, sense RNAs in blue. Black triangles denote promoters. Light blue, ribosome binding sites (RBS). Yellow symbols indicate ribosomes. Green arrows denote RNase III cleavage; black arrows indicate unknown RNase action. The violet symbol represents RNase R. For details, see text. B, C, E, F, and H are based on reference 13.
Table 2. Overview of cis-encoded sRNAs from low-GC Gram-positive bacteria.
| Antisense RNA/Target-RNA | Length of sRNA (nt) | Species | Biological function | Mechanism | Specific characteristic |
|---|---|---|---|---|---|
| RatA/txpA | 222 | B. subtilis | antitoxin/toxin | RD | |
| SR4/bsrG | 180 | B. subtilis | antitoxin/toxin | RD + TI | bsrG temperature dependent |
| SR5/bsrE | 163b | B. subtilis | antitoxin/toxin | ? | |
| antibsrH/bsrHc | ≈202 | B. subtilis | antitoxin/toxinc | ? | |
| antiYonT/yonTc | ≈80 | B. subtilis | antitoxin/toxinc | ? | |
| ECF/yab | 750 | B. subtilis | autolysin | RD* | σX, σM |
| SprA1AS/SprA1 | 60 | S. aureus | antitoxin/toxin | RD | SprA1 is additionally a cytolisin and acts on human erythrocytes |
| RsaOX/sa0062* | 129 | S. aureus | transposase? | RD* | |
| RsaOW/is1181 | S. aureus | transposase? | ? | 8 RsaOW copies | |
| p3 RNA/glnA | 43 | C. acetobutylicum | N metabolism | ? | N-induced (p3) |
| Antisense/ubiG | 264/424/730/1000 | C. acetobutylicum | S metabolism | TINF | S-box riboswitch |
a Renamed (Jahn and Brantl, unpublished). bLength determined (Maiwald, Jahn, Brantl, unpublished). cNot yet demonstrated in Bacillus that indeed TA system. RD, RNA degradation; TI, translation inhibition; TINF, transcriptional interference; *, mechanism proposed but not experimentally substantiated; ?, no mechanism proposed.
Table 3. Overview of trans-encoded antisense RNAs from low GC Gram-positive bacteria.
| sRNA (nt) | target RNA(s) | Biological function | Mechanism of action | Control of expression/ specific characteristic |
|---|---|---|---|---|
| Bacillus subtilis | ||||
| SR1 (205) | ahrC | arginine catabolism | TI | CcpN, CcpA |
| gapA operon* | sugar metabolism | SR1P/GapA* | ||
| FsrA (84) | sdhCAB, citB | succinate dehydrogenase | TI | Fur, iron; some targets need FbpA, B, or C |
| gltA | glutamate synthase (iron-sulfur) | |||
| lutABC | iron-sulfur oxidase | |||
| dctP | dicarboxylate permease | |||
| yvfW, leuCD | ||||
| RsaE (114) | cstAa, sucCa, galKa central metabolisma | TI | exponential phase | |
| Staphylococcus aureus | ||||
| RNAIII (514) | hla | α hemolysin | TA | AgrC/AgrA |
| rot | repressor of toxins Host-pathogen interactions |
TIb and RD | stationary phase | |
| sa1000/sa2261 | fibrinogen BP, ABC transporter | |||
| spa | protein A | |||
| coa | coagulase | |||
| lytM | peptidoglycan hydrolase | |||
| map | MCH class II analogous protein | TA? | ||
| RsaE (96) | opp-3B/3A, sucC, sA0873 | central metabolism | TI | pre-stationary phase, heat-shock |
| SprA (202) | sa2216# | ABC transporter? | Post-translational? | strain-specific |
| SprD (142) | sbi | immune response | TI, RD | growth phase |
| Psm-mec (157) | agrA | virulence regulator | TI, RD independent of TI | |
| Art A (345) | sarT | transcriptional regulator of hla | RD | ArgA |
| Streptococcus pyogenes | ||||
| Pel RNA (459) | speB, | cysteine protease | post-transcriptional | Multiple transcription regulators and conditioned media |
| emm, sic, nga | M- and -related proteins | transcriptional control | ||
| FasX (205) | ska | streptokinase | RNA stabilization | FasBCA, luxS |
| sagS | streptolysin | ? | aa starvation | |
| fbp54 | fibronectin binding protein | ? | ||
| mrp | fibrogene binding protein | ? | ||
| RivX (289,237,189) | mga | virulence | TA? | CovR/CovS |
| tracrRNA (171,89) | crRNA precursor | CRISPR maturation | RNA processing | Csn1b |
| S. pneumoniae | ||||
| srn206 (120) | comD | sensor histidine kinase, competence | TI? | |
| csRNA1–5 (87–151) |
comC | competence | TI? | All 5 homologous sRNAs are regulated by CiaRH |
| spr0081 | ABC transporter | |||
| spr0159 | DNA binding protein | |||
| spr0551 | branched chain aa transport | |||
| spr1097 | formate/nitrite transporter | |||
| Clostridium perfringens | ||||
| VR (386) | pfoA, vrr, virT, virU, ccp | collagenase, toxin genes | RNA processing | VirR/VirS |
| CE1446/CE1447 | TA? | |||
| VirX(?) | pfoA, plc, colA | α, κ, τ enterotoxin production | TA? | independent of VirR/VirS |
| spo0A σE, σF, σK | sporulation | encodes 51 aa peptide VirX | ||
| Listeria monocytogenes | ||||
| LhrA (268) | lmo0850 | protein of unknown function | TI and RD | Hfq required for stability and target binding |
| chiA | two chitinases | TI | ||
| lmo0302 | protein of unknown function | TI | ||
| SreA (228) | prfA | virulence master regulator | TI | SAM RS, blood |
| lmo0049 = agrD | quorum sensing molecule | TA? | ||
| SreB (179) | prfA, lmo2230 | arsenate reductase homol. | ? | |
| RilB | lmo2104/2105# | iron transport proteins | ? | |
| RilE | comEA/FA, lmo0945# | competence factors | ? | |
| RliI | lmo1035# | phosphotransferase system | ? | |
| Rli23, 25, 35 | lmo0172# | transposases | ||
| Rli45 | rli46# | ? | ||
| Rli29 | lmo9471# | ? | ||
| Rli30 | lmo0506# | ? | ||
Only sRNAs are listed for which target genes have been identified or #proposed. aIn analogy to S. aureus RsaE, which interacts via C-rich loops with SD sequences, targets have been predicted. bRequired for processing of the sRNA/target RNA duplex by RNase III *SR1 acts on gapA as a peptide encoding mRNA, i.e., SR1P interacts with GapA, thereby stabilizing gapA mRNA. TI, translation inhibition; TA, translation activation; RD, mRNA degradation. Control of expression: All proteins and growth conditions known to regulate sRNA expression are listed. It is not indicated whether these factors promote or inhibit sRNA expression. For more details, see text. RS, riboswitch.
Chromosome-Encoded sRNAs in Low GC Gram-Positive Bacteria and Their Biological Functions
In the past years, a variety of articles have been published on the identification of sRNAs in Gram-positive bacteria (reviewed in ref. 13). On average, ≈100–200 sRNAs have been discovered in a single genome. Despite newly available methods, it is still a challenging task to identify the targets of these novel riboregulators. In the following, we focus on base-pairing sRNAs for which targets have been verified experimentally. For an overview that also includes data on riboswitches and protein-binding sRNAs from Gram-positive bacteria, see reference 14.
Chromosome-encoded sRNAs are involved in a wide variety of biological functions. Mostly, they fine-tune metabolic processes and regulate stress adaptation or virulence. Fine-tuning functions are reflected by the lack of severe phenotypes upon deletion or overexpression of such RNAs. Examples for metabolic regulation include arginine catabolism (B. subtilis SR115) and iron-transport and storage (B. subtilis FsrA16) or central metabolism (S. aureus RsaE17).
A few trans-encoded sRNAs contain additionally small open reading frames (ORFs) that are translated. Such RNAs were designated dual-function sRNAs. Examples for small ORFs are the 26 codon δ-hemolysin ORF of S. aureus RNAIII,18 the 22 codon psmα-ORF in S. aureus Psm-mec,19 the streptolysin SLS-ORF of Streptococcus Pel RNA,20 and the 39 codon ORF sr1p on B. subtilis SR1 RNA (see below21). The translation products of the small ORFs can either operate in the same pathway as the base-pairing sRNA (in RNAIII and Pel RNA) or in another pathway: B. subtilis SR1 acts as a base-pairing sRNA in arginine catabolism, whereas SR1P acts in sugar metabolism.21 To date, neither for the 72 codon hyp7 ORF of Clostridium perfringens VR22 nor the 32 codon ORF of Streptococcus pyogenes RivX23 data are available on translation or possible biological function(s).
In different approaches, genome-wide overlapping (antisense) transcription was found (reviewed in ref. 24). Antisense transcription in the same bacterial cells yields a collection of short RNA fragments that result from RNase III processing, which appears most prominently in Gram-positive bacteria. Examples are provided below for B. subtilis and S. aureus. The mechanisms through which overlapping transcription can affect sense RNA expression are diverse. Primarily direct base-pairing interactions between sense and antisense transcripts, which result in RNase III cleavage of the complexes, can be imagined. An alternative is transcriptional interference that has, so far, been documented only in one case (see below).
In the following, sRNAs discovered in Bacillus subtilis, Staphylococcus aureus, Listeria monocytogenes, as well as Clostridium and Streptococcus species, and for which targets have been identified, are discussed in detail with regard to their biological function, expression conditions, and mechanism of action.
sRNAs from Bacillus subtilis
Trans-encoded sRNAs
The first trans-encoded sRNA in B. subtilis, FsrA (84 nt) was discovered in 200816 and resembles E. coli RyhB as it is transcriptionally repressed by Fur and regulates target mRNAs involved in iron metabolism and storage, e.g., sdhCAB (succinate dehydrogenase) and citB (aconitase). Using transcriptomics, additional FsrA targets were identified, among them, gltAB encoding the iron-sulfur-containing enzyme glutamate synthase and lutABC, dctP, resA, and qcrA.25 Consequently, FsrA is a global regulator in B. subtilis. In contrast to E. coli RyhB, which requires the RNA chaperone Hfq, FsrA cooperates with one, two, or three Fur-regulated small basic proteins, FbpA, FbpB, and FbpC, suggested to be RNA chaperones to repress translation of its different targets.16 Under iron-limited growth conditions, both FsrA and FbpB (48 aa) inhibit translation of lutABC encoding iron-sulfur-containing oxidases, which allows iron to be directed to higher-priority target proteins.26 FsrA is predicted to target the region upstream of and including the RBS, while FbpB might facilitate FsrA/lutABC RNA pairing or recruit the RNA degradation machinery.26 Thereby, FsrA is the main regulator, as its modest overexpression can bypass the need for FbpB.26 The authors suggest that FsrA uses a C-rich single-stranded region to interact with the ribosome binding sites of its different target mRNAs.25
In 2009, it has been found that the expression of BsrF (115 nt) is controlled by the global regulator CodY that responds to branched chain amino acids and GTP.27 However, no BsrF targets have been identified so far. In 2011, CsfG, a sporulation-specific, non-coding sRNA highly conserved in endospore formers was found.28 Its target has not yet been determined. The same holds true for a number of non-coding sRNAs that are under sporulation control, which were discovered in 200629 and further analyzed later on.30 Among them, SurC is transcribed under control of σK and is conserved in the distantly related B. anthracis.29
SR1—The first dual-function sRNA in Bacillus subtilis
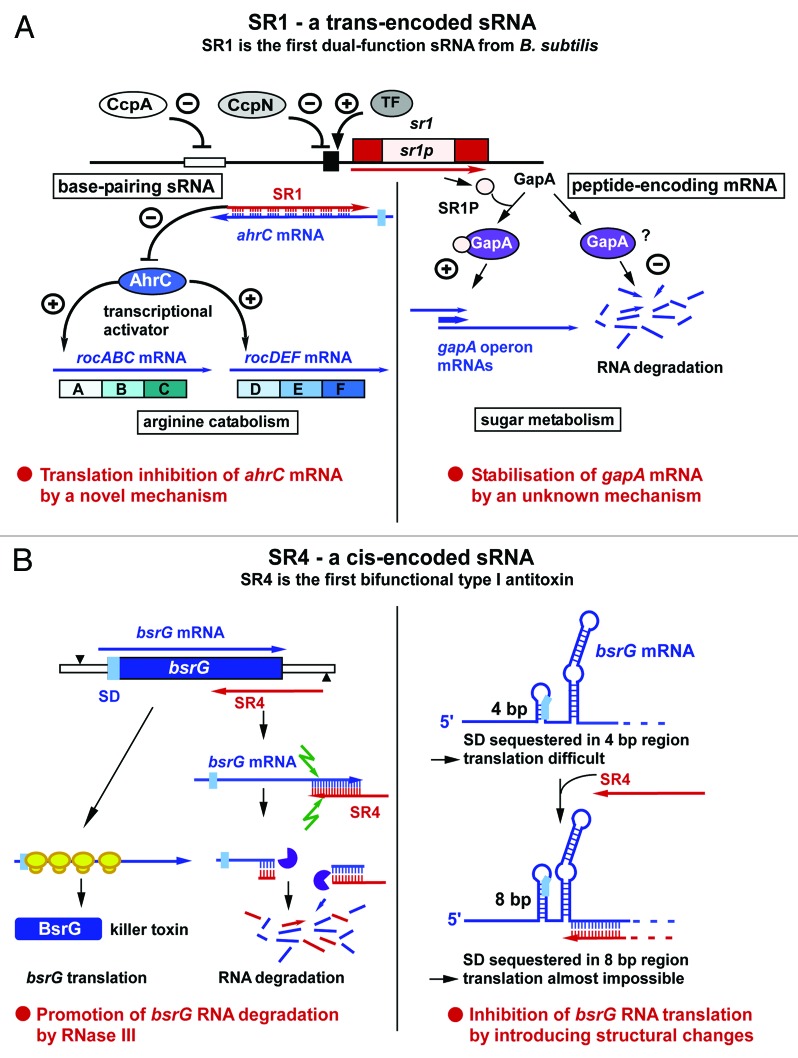
The small RNA SR1 was discovered using a bioinformatics approach for sRNAs in intergenic regions of the B. subtilis chromosome combined with northern blotting.15 SR1 comprises 205 nt and is encoded between pdhD and speA. Knockout and overexpression of sr1 were not detrimental for B. subtilis growth. Using 2D gel electrophoresis and northern blotting with a wild-type and an sr1-knockout strain, two arginine catabolic operons—rocABC and rocDEDF—were identified as secondary targets whose expression is upregulated in an sr1-knockout strain. The mRNA encoding AhrC, the transcription activator of these operons, is the primary target of SR1.31 Both RNAs share seven regions of complementarity in the central part of ahrC and the 3′ half of SR1 (Fig. 2A). SR1 does not affect the stability or amount of ahrC mRNA, but inhibits its translation by a novel mechanism32: Although it binds ≈100 nt downstream of the ahrC RBS, it induces structural changes 20–40 nt downstream of the ahrC RBS that inhibit translation initiation (Fig. 1B). This was shown by secondary structure probing of the ahrC/SR1 complex and toe-printing studies.32 SR1 is only expressed under gluconeogenic conditions and 20–30-fold repressed by CcpN, and, to a minor (3-fold) extent, CcpA, under glycolytic conditions.15 CcpN binds upstream of and overlapping the sr1 promoter,33 whereas CcpA binds 250 upstream of the transcription start site (TSS) at a cre site. CcpN represses sr1 transcription in the presence of ATP and slightly acidic pH (6.5)34 by interacting with the α-subunit of the RNA polymerase, thereby inhibiting promoter escape.35 Transcriptomics and northern blotting suggested a second target for SR1, the gapA operon. In the presence of SR1, gapA mRNA is stabilized, whereas it is barely detectable in the absence of SR1 under gluconeogenic conditions. We demonstrated that the 39 aa peptide encoded by SR1, SR1P, is responsible for this effect: SR1P binds to GapA (glyceraldehyde-3P dehydrogenase A), thereby stabilizing gapA operon mRNA by a hitherto unknown mechanism.21 Consequently, SR1 is a dual-function sRNA: It acts as a base-pairing regulatory RNA on ahrC RNA, and as a peptide-encoding mRNA on the gapA operon.
Figure 2. SR1 and SR4, a trans- and a cis-encoded sRNA from B. subtilis. As in Figure 1, the antisense RNAs are indicated in red, the sense RNAs in blue, RBS in light blue, ribosomes are in yellow, RNase III in green, and RNase R in violet. (A) SR1, a trans-encoded sRNA, is the first identified dual-function sRNA from B. subtilis. +, activation; -, repression. CcpA and CcpN repress sr1 transcription under glycolytic conditions. TF is a novel transcription factor that activates sr1 transcription at cold-shock. (B) SR4, a cis-encoded sRNA, is the antitoxin of the type I TA system bsrG/SR4. It is the first antitoxin for which two independent functions have been found. For details, see text.
Recently, a computer prediction revealed SR1 homologs in 23 other species belonging to the Bacillales.36 The expression of the SR1 homologs in seven of these species was experimentally verified. Furthermore, the ability of SR1P homologs to bind B. subtilis GapA was investigated in northern blots and co-elution experiments, and the interaction between SR1 homologs and the corresponding ahrC homologs studied in vitro.36 The results demonstrated that both functions of SR1, the base-pairing and the peptide-encoding function, are remarkably conserved over ≈1 billion of years of evolution. Figure 2A represents the two functions of SR1.
To elucidate the interaction surface of SR1P and GapA, a series of peptide mutants were constructed and analyzed in northern blotting and co-elution experiments. To verify the predicted binding regions, both protein-coding genes have to be mutated and expressed simultaneously and independently in B. subtilis. For this purpose, the pMG vector family, a series of modular plasmids suitable for chromosomal integration and gene expression under single copy conditions in B. subtilis, was constructed.37 Preliminary data show that for the stabilizing effect of GapA/SR1P, only part of the gapA operon mRNA must be present, which must contain sequences adjacent to the gapA ORF (Gimpel, unpublished).
Recently, we observed that the amount of SR1 is significantly reduced after an 18 °C cold-shock. This was due to altered transcription initiation and not RNA degradation. Upon investigation of the cold-shock effect, we discovered a novel trigger enzyme (Preis et al., unpublished).
Future work will be aimed at the investigation of the molecular mechanism behind the SR1P–GapA interaction, in particular, the determination of the SR1P binding pocket and properties as KD-value or stoichiometry of the interaction. Moreover, the biological role of SR1P-GapA interaction will be elucidated.
Cis-encoded sRNAs
With the exception of ECF RNA (see below), only cis-encoded sRNAs that act as type I antitoxins have been identified so far in B. subtilis. Currently, five types of toxin-antitoxin systems (TA systems) are known (reviewed in ref. 38). In type I TA systems, the antitoxin is a small RNA, and the toxin mRNA encodes a hydrophobic peptide. RNA antitoxins are cis-encoded regulatory RNAs that interact with their target RNAs either at their 5′ or 3′ end by a base-pairing mechanism. In Bacillus subtilis, 14 type I TA systems have been postulated, and three of them verified experimentally: txpA/RatA,39 bsrG/SR4,40 and yonT/as-yonT.41,42 The majority of them are located on prophage elements or phage remnants in the chromosome.
RatA—The first identified RNA antitoxin from B. subtilis
In 2005, txpA/RatA was identified as the first type I TA system on the skin element of the B. subtilis chromosome.39 In the absence of RatA, TxpA causes cell lysis on agar plates. The RatA RNA is 220 nt long and overlaps the 3′ end of txpA mRNA by ≈120 nt.39 The interaction between txpA mRNA and RatA promotes the degradation of txpA mRNA by an RNase III-dependent mechanism.42 Degradation of txpA mRNA by RNase III is essential for viability of B. subtilis.42 The secondary structures of RatA and txpA RNA, as well as their complex, have been determined.42 The ribosome binding site (RBS) of txpA is located in a 5 bp ds region. This sequestration is not altered upon RatA binding.
SR4—An RNA antitoxin with two functions
The bsrG/sr4 module is located on the SPβ prophage of the B. subtilis chromosome. We corroborated experimentally that this module acts as a type I toxin/antitoxin system:40 The bsrG RNA is 294 nt long and codes for a 38 aa toxic peptide with a predicted central transmembrane domain and a charged C terminus (see ref. 41). The antisense RNA SR4 is 180 nucleotides long and overlaps the 3′ end of bsrG by 123 nucleotides. The interaction of both RNAs at their 3′ ends promotes degradation of bsrG RNA. RNase III cleaves the bsrG RNA/SR4 duplex at position 185 of bsrG RNA, 8 nt downstream from the stop codon, but is not involved in the degradation of either bsrG RNA or SR4 alone.40 Endonuclease Y and the 3′-5′ exoribonuclease R are responsible for further degradation of both RNAs. PNPase processes SR4 precursors into the mature RNA. As a Δrnc suppressor strain neither lysed on agar plates nor had mutations in the bsrG ORF, RNase III is not essential to the function of the bsrG/SR4 system.40 Later it was found that the Δrnc suppressor strain has a deletion of the skin prophage and a tendency to loose SPβ in order to reduce TxpA and YonT toxicity.41 Hfq is not required for the function of the bsrG/SR4 system,40 since a Δhfq strain does neither show lysis nor altered half-lives of bsrG RNA or SR4.
Recently, the secondary structures of SR4 and bsrG-RNA, as well as the SR4/bsrG RNA complex, were experimentally determined. The results demonstrated that SR4 induces structural alterations around the RBS of bsrG43 (Fig. 2B): A 4 bp region that sequesters the bsrG RBS is extended to 8 bp in the presence of SR4. It was shown experimentally that this extended double-stranded region inhibits translation. Consequently, SR4 is the first type I antitoxin with two clearly separable functions: it promotes degradation of bsrG mRNA and impedes ribosome access to the bsrG SD, thereby preventing translation. Complex formation assays with wild-type bsrG RNA/SR4 yielded an apparent binding constant kapp of 6.5 × 105 M−1 s−1,43 which is in the same order of magnitude as those of other cis-encoded sense/antisense RNA pairs.44 The binding pathway of bsrG mRNA and SR4 was elucidated: Binding starts with a single loop–loop contact between loop 3 of bsrG RNA and loop 4 of the SR4 terminator stem-loop. Intermolecular base-pairing progresses via the single-stranded region between L4 and L3 toward loop 3 of SR4, and, finally to loop 2, which pairs with the bsrG terminator-stem-loop. However, the latter interaction was not required for efficient binding.43 Loop 3 of bsrG RNA contains a 5′ YUNR motif, which apparently forms a U-turn that provides a scaffold for the efficient initial interaction between both RNAs.43 The sr4 promoter is about 6- to 10-fold stronger than the bsrG promoter,40 which should result in an excess of the antitoxin over the toxin, as was shown for txpA/RatA.42
Other cis-encoded sRNAs in B. subtilis
A recent review summarizes the current knowledge about other cis-encoded sRNAs that act as type I antitoxins in B. subtilis.41 Among them is yonT80, which is regulated by as-yonT, and was recently confirmed to be a toxin in E. coli, and indirectly in B. subtilis.42 Furthermore, BsrE, regulated by as-BsrE (renamed SR5) acts as a toxin in B. subtilis (Maiwald, Jahn, and Brantl, unpublished).
The only other cis-encoded sRNA known to date is the long (750 nt) ECF RNA, which is expressed under control of extracytoplasmic sigma factors σX and σM and was found to regulate an autolysin encoded by the yabE gene.45
A transcriptome analysis indicated a widespread antisense transcription in bacterial chromosomes: Using micorarrays, for 2.9% of all B. subtilis genes, antisense transcripts were detected,2 and a dRNA Seq approach found in total 29 cis-encoded antisense RNAs.3 To date, the biological role of these antisense RNAs is unknown.
In Staphylococcus, Streptococcus, Listeria, and Clostridium species, mostly sRNAs implicated in the regulation of pathogenesis and virulence, but also a few metabolic riboregulators, have been discovered and investigated in more detail.
sRNAs from Staphylococcus aureus
Meanwhile, six different approaches have identified about 100 trans- and 100 cis-encoded sRNAs in the human pathogen S. aureus (Table 1). The majority of the hitherto characterized sRNAs are involved in pathogenesis. The most prominent and best-characterized of them, which was already discovered in 1995, is the unusually long RNAIII (514 nt).18 Transcription of RNAIII is induced by AgrA, the response regulator of the quorum-sensing agr (accessory gene regulation) system, and RNAIII is the effector of this system. Its secondary structure was mapped in vitro and in vivo and revealed 14 hairpin structures, out of which the conserved hairpin 13 was involved in repression of protein A expression.46 RNAIII was the first antisense RNA for which an activating function has been found18 (Fig. 1C). However, it does not only activate translation of the α-hemolysin mRNA, but also inhibits translation of a variety of targets:47 spa (main surface adhesin protein), SA1000 (novel fibrinogen-binding protein), sa2261 (ABC transporter), rot (pleiotropic transcriptional factor Rot48), lytM (peptidoglycan hydrolase), and coagulase mRNA.49 To exert its inhibitory effect, RNAIII employs a combination of translation inhibition by a base-pairing interaction of its two redundant 3′ hairpin loops with the target mRNA and recruitment of RNase III for target degradation.50 As shown for spa, RNA degradation was required for permanent translational arrest. Specificity for RNAIII on all translationally inhibited targets is obtained by either propagating the first loop–loop contact at the RBS into the stem regions (sa1000 and sa2353 mRNAs) or by addition of a second loop–loop interaction (rot and coa mRNAs). The RNAIII/coa mRNA duplex comprises an imperfect duplex masking the SD sequence and a loop–loop interaction in the coa ORF. The imperfect duplex is sufficient to prevent translation initiation. RNase III cleaves the two regions of the coa mRNA bound to RNAIII that may contribute to the degradation of the repressed mRNA.49 Interestingly, RNAIII represents the first identified dual-function sRNA as it is on the one hand a regulatory sRNA that acts by base-pairing and on the other hand a protein-encoding mRNA with an ORF for the 26 aa δ-hemolysin.18
The second dual-function sRNA in S. aureus, Psm-mec (157 nt), was discovered only in 2013.19 The psm-mec gene is located on the mobile genetic element SCCmec that confers—via mecA—methicillin resistance to MRSA strains. Psm-mec encodes the 22 aa-secreted cytolytic toxin PSMα (α phenol-soluble modulin). Additionally, it inhibits translation of agrA mRNA by base-pairing of its 5′ nt 21–50 with the agrA coding sequence (most important are nt 199 to nt 267). Furthermore, it causes a ≈2-fold RNase III-dependent decrease in agrA half-life, which was—in contrast to the inhibitory effect of RNAIII on rot mRNA—independent of the translation inhibition effect. Psm-mec RNA itself is stable (half-life 20 min) and not affected by agrA mRNA. AgrA activates transcription of RNAIII and of the psmα 3 operon. Twenty-five percent of 325 analyzed clinical isolates (HA strains) have a promoter mutation that causes attenuated psm-mec transcription, and 9% have no psm-mec, which results in high virulence. By contrast, community-acquired MRSA strains (CA strains) have no Psm-mec. Kaito et al. proposed that Psm-mec sRNA attenuates virulence in HA comparison to CA strains.19
Recently, a second AgrA-regulated sRNA, ArtA, was discovered that controls α-toxin expression by targeting the 5′ UTR of sarT mRNA.51
In 2005, a search in the clinical agr negative S. aureus strain N315 resulted in seven experimentally confirmed small pathogenicity islands RNAs = Spr (SprA to SprG), among which SprA, E, F, and G were present in multiple copies, partly also on the core genome.52 For SprA (202 nt), in vitro data suggested an interaction with three putative mRNA targets, among them a 3.5 kb ABC transporter mRNA.52 However, in 2012 it was found instead that SprA1 encodes a 30 aa peptide toxin, which inserts into the cell membrane and kills S. aureus cells, and is regulated by a cis-encoded antisense RNA (SprA1AS) that facilitates degradation of SprA1 mRNA. Therefore, SprA1/SprA1AS constitutes a type I toxin-antitoxin system.53 SprA1AS combines—similar to RNAI from E. faecalis plasmid pAD1 (reviewed in ref. 13)—features of a cis- and a trans-encoded sRNA.54 Surprisingly, the SprA1 peptide was previously also found to act as cytolysin on human erythrocytes.54 In 2010, SprD (142 nt), another sRNA of the first search, was shown to inhibit translation initiation of the abundant secreted immune-evasion protein Sbi by an interaction between its central region and the 5′ 41 nt of sbi mRNA, including RBS and start codon.55 In contrast to RNAIII and Psm-mec, SprD did not affect the sbi mRNA levels. In an animal septicaemia model, SprD impaired both adaptive and innate host immune responses.54
The first staphylococcal sRNA involved in metabolic regulation, RsaE, was discovered in 200917 among 11 novel Hfq-independent sRNAs (RsaA–RsaK). RsaE (96 nt) is highly conserved in four differential S. aureus species and was also found in Macrococcus and Bacillus.17 It co-regulates several metabolic pathways involved in amino acid and peptide transport, cofactor synthesis, carbohydrate and folate metabolism, and TCA cycle by inhibiting translation of two cistrons of an oligopeptide transporter operon, opp-3A56 and opp-3B,17 and sucC/sucD-encoding succinyl-CoA synthetase subunits α and β, and by targeting sA0873.56 Expression of rsaE is AgrA-dependent and very low in pre-stationary phase. Similar to B. subtilis FsrA and S. aureus RNAIII, RsaE seems to recognize its target mRNAs at the RBS via a conserved C-rich loop (UCCC motif). In another screen for sRNAs, RsaOX (129 nt) and RsaOW were proposed to target the transposase genes sa0062 and is1181, respectively, possibly by promoting RNA degradation.56
Recent tiling arrays revealed long antisense RNAs that are rapidly degraded by RNase III.57 It is suggested that such long sRNAs might play an important role in staphylococcal gene regulation, in particular, of genes involved in pathogenesis and virulence. One example is SSR42 (891 nt) that affects erythrocyte lysis and pathogenesis in a murine infection model.58 SSR42 is stabilized in stationary phase, upregulates genes involved in capsule biosynthesis, and downregulates ≈80 genes, among them, virulence factors. However, its role seems to be indirect as no SSR42 binding to the corresponding mRNAs was found.58 A summary of all kinds of regulatory RNAs hitherto found and investigated in Staphylococcus areus and their biological functions has been published recently.59
sRNAs from Clostridium
In 2013, RNA-seq approaches in three Clostridium species discovered between 36 and 182 sRNAs (Table 1). So far, for only a few of them, targets are known. Already > 20 y ago, a cis-encoded antisense RNA was found in the biotechnologically important Clostridium acetobutylicum, which is involved in control of nitrogen metabolism by interacting with the 5′ UTR of the glutamin synthetase gene glnA.60,61 However, no further reports were published on this issue. In the same species, four antisense RNAs were discovered in 2008 that are encoded downstream of the ubiG operon and act in concert with an S-box riboswitch to regulate sulfur metabolism,62 Figure 1F. These long antisense RNAs (between 200–1000 nt) represent the so-far-unique example for transcriptional interference as mechanism of action of base-pairing sRNAs. In the human pathogen Clostridium difficile, a genome-wide association study identified 94 trans-encoded sRNAs and 91 cis-encoded sRNAs, and confirmed 35 of them experimentally.7
In the food-born pathogen Clostridium perfringens, two sRNAs have been characterized in more detail: The sRNA VR is part of the VirR/S regulon that controls toxin production and induces collagenase (K-toxin) and b2-toxin synthesis,63 Figure 1F. VR has been shown to regulate five genes by direct binding to their mRNAs: pfoA, vrr, virT, ccp, and virU. Additionally, VR positively affects synthesis of CPE1447 and CPE1446, which form a protein heterodimer that controls toxin gene expression.64 The function of the small ORF encoded on VR is still unclear. In 2013, another sRNA, VirX, which had been shown before65 to regulate pfoA, plc, and colA mRNAs independent of the VirR/VirS system has been found to repress genes encoding positive sporulation regulators like Spo0A and sigma factors E, F, and K.66 Inactivation of virX led to higher levels of sporulation and enterotoxin production. Data on sRNAs in Clostridium species available until 2011 have been summarized.67
sRNAs from Listeria
Although five different approaches have discovered sRNAs in Listeria monocytogenes and L. innocua (Table 1), only a few targets have been identified so far. One screen for Hfq-binding sRNAs in L. monocytogenes identified LhrA, LhrB, and LhrC.68 LhrA is stabilized by Hfq and targets at least three mRNAs directly, chiA mRNA encoding two chitinases and two genes of unknown function (lmo0850 and lmo0302).69,70 In a global screen, 300 genes were found to be affected by LhrA. In the presence of Hfq, LhrA inhibits translation of chiA mRNA (Fig. 1A). LhrA is, so far, the only example from Gram-positive bacteria for which an effect of the RNA chaperone Hfq on target binding was found69 (see below). However, the majority of the sRNAs from Listeria do not seem to need Hfq for stabilization or interaction with their targets (reviewed in ref. 69). In the case of lmo0850, LhrA both inhibits translation and promotes RNA degradation. For a few recently discovered sRNAs, targets were predicted, but their mechanism of action on them is still elusive71: RliB is proposed to target lmo2104/5 involved in iron transport, RliE the competence factors comEA/FA, and RliL a phosphotransferase system (lmo1035). The absence of 15 of the 29 sRNAs recently found in L. monocytogenes72 in the non-pathogenic L. innocua underlines the importance of riboregulators for pathogenesis and virulence. Rli38 from L. monocytogenes is 25-fold higher expressed in blood and in the presence of H2O2, i.e., under oxygen stress.
In RNaseq approaches, unusually long antisense RNAs (las RNA) complementary to more than one ORF or operon were found in Listeria species. The authors designated such a genomic locus excludon.72,73 Thereby, the 5′ or 3′ non-coding part of a lasRNA negatively affects the expression of one or several gene(s) on the complementary strand, whereas the remaining (major) part functions as mRNA for the downstream or upstream genes. The first reported excludon in L. monocytogenes controls flagellum biosynthesis at the motility gene repressor locus. MogR is the transcriptional repressor of flagellum and motility genes in Listeria species, and mogR is transcribed from a promoter 45 nt upstream of the AUG. In opposite direction, the flagellum operon with lmo0675 (unknown), lmo0676 (fli), lmo0677 (fliQ), and lmo0678 (fliR) is transcribed. FliP and FliQ form the flagellum export apparatus. Additionally, transcription initiated at a third promoter upstream of pmogR results in an excludon transcript, Anti0677, whose 5′ region is antisense to lmo0675-lmo0677, and whose 3′ part contains the mogR ORF. Anti0677 is under control of stress-activated σB located within the lmo0677 ORF. Other recently summarized examples73 comprise two putative permease-efflux pump excludons and a putative carbon source utilization excludon. However, the detailed mechanism of action of the proposed lasRNAs has not yet been elucidated.
In 2009, two trans-acting S-adenosylmethionine riboswitches (SreA and SreB) that can function as trans-encoded sRNAs were discovered in L. monocytogenes.74 SreA upregulates argD and represses translation of prfA encoding the virulence master regulator by base-pairing upstream of the RBS. SreB (179 nt) regulates prfA and lmo2230 (arsenate reductase homolog).74 By targeting PrfA, SreA and SreB link virulence to nutrient availability.
sRNAs from Streptococcus
Streptococci include species that cause severe human diseases. In the group A (GAS) Streptococcus pyogenes, 75 sRNAs were identified (Table 1), and three of them were implicated in virulence control: Pel, FasX and RivX. The 450 nt long bifunctional sRNA Pel regulates M- and M-related proteins and codes for streptolysin S (SLS).20 It exerts pleiotropic effects on virulence. Pel expression is repressed both by a multitude of transcription factors, in fact CcpA, CovRS/CsRS, LuxS, Mga, Nra, and RopB/Rgg, and, additionally, by FasX RNA, but activated at amino acid starvation by CodY, Irr, and SLS itself.
FasX (≈200 nt) is encoded in the fasBCAX operon and transcriptionally activated by response regulator FasA. FasX stabilizes the ska mRNA encoding the secreted plasminogen activator streptokinase75 (Fig. 1E). It also controls sagS mRNA encoding streptolysin S (see above) and, by unknown mechanisms, transcription of fbp54 mRNA and mrp mRNA encoding two fibronectin-binding proteins. In S. dysgalactiae ssp. equisimilis (group C streptococci GCS), FasX also affects ska and streptolysin.76
The third recently found sRNA, RivX, is processed from a longer mRNA-encoding transcription regulator RivR and has, thus, three alternative 5′ ends resulting in 189, 237, 289 nt-long species. Together with transcription factor RivR, RivX upregulates the global virulence gene regulator mgA by enhancing its translation.23 Mga itself controls emm, C5a peptidase, and cysteine protease speB. As mentioned above, RivX contains a 23 codon ORF of unknown function.
In 2011, tracrRNA (89 nt), a trans-encoded sRNA involved in maturation of CRISPR RNA was discovered.12 CRISPR protects its host against prophage-derived DNA. In type II CRISPR systems, tracrRNA induces—together with Cas9 and RNase III—cleavage of pre-crRNA to yield mature crRNA (Fig. 1G), which, upon phage infection, can target phage DNA. By probing selected loci, functional tracrRNA homologs were also found in Streptococcus mutans, S. thermophiles, and Listeria innocua, and even the Gram-negative Neisseria meningitidis.12
Details about expression regulation and the involvement of streptococcal sRNAs in global networks controlling virulence and pathogenesis have been summarized recently.77
sRNAs from Streptococcus pneumoniae
In Streptococcus pneumoniae, far more than one hundred sRNAs have been identified mainly by high-throughput approaches (Table 1). One sRNA, srn206, has been implicated in competence control78 and the target was suggested to be comD, the gene encoding the histidine kinase of the two-component regulatory system ComDE, which is essential for initiation of competence development.79 Several sRNAs were shown to be involved in the control of various aspects of virulence and a number of differentially expressed proteins were detected in sRNA mutants.80 However, no direct regulatory link was established between sRNAs and putative targets. In addition, two-component regulatory systems appeared to be involved in sRNA expression control, but the underlying mechanism was not determined.80 Despite the identification of numerous sRNAs in whole genome approaches, no clearly defined targets or regulatory mechanisms were determined so far.
More information is available for the first sRNAs described in S. pneumoniae, which have been detected in an analysis to define the regulon of the two-component regulatory system CiaRH.81 The five strongest promoters in the CiaRH regulon were found to drive expression of sRNAs between 87–151 nt in size. These non-coding sRNAs, designated csRNAs (cia-dependent small RNA), show a high degree of similarity to each other. They are predicted to adopt a secondary structure with two stem-loops at the 5′- and 3′-ends, respectively. Sequences complementary to the SD sequence and the start codon AUG are present in the unpaired region between the stem-loops. The csRNAs appear to affect pneumococcal physiology pleiotropically. Stationary phase autolysis was affected by csRNA4 and csRNA5,81 and a csRNA5 mutant was defective in lung infection.80 Furthermore, csRNA1 was shown to act negatively on competence development.82 The csRNAs were originally detected in S. pneumoniae R6,81 but are found in all S. pneumoniae genome sequences available to date. Curiously, Hungarian S. pneumoniae serotype 19A isolates carry and express longer versions of csRNA5, which apparently arose by internal sequence duplication (R. Brückner, unpublished observations). Expression of CiaR-controlled csRNAs was also confirmed in Streptococcus mitis, Streptococcus oralis, Streptococcus sanguinis,83 and Streptococcus pyogenes.84 The presence of multiple csRNAs genes could be predicted in all streptococcal genomes83 suggesting that they serve an important function in this group of organisms.
In a recent study, csRNA target predictions in S. pneumoniae R6 were evaluated by analyzing translational fusions of candidate genes.85 Six targets could be identified, which were all downregulated by the csRNAs. At least for the three genes tested, each of the csRNAs could act upon the targets reflecting the high degree of csRNA similarity. Regulation by the csRNAs was additive, no single csRNA was as effective as all csRNAs together. Four of the regulated genes, spr0081, spr0371, spr0551, spr1097, encode transport proteins of various protein families, but their physiological roles in S. pneumoniae are currently unknown. A putative transcriptional regulator spr0159 and comC, the gene encoding the precursor of the competence stimulating peptide CSP86 were the remaining csRNA targets. Especially the identification of the latter was intriguing, since CiaRH was known to act negatively on competence development.87,88 Mutation of comC between the SD sequence and the start codon partially disrupting complementarity to the csRNAs greatly diminished csRNA-mediated repression of a translational fusion. Replacing wild-type comC by the mutated version in the genome of S. pneumoniae R6 relieved competence from CiaRH-dependent control.85 Therefore, the csRNAs block CSP precursor production thereby interfering with pheromone signaling that initiates competence.
Interestingly, CiaRH controls production of the serine protease HtrA, which is also able to act negatively on competence by degradation of CSP.89 Which negative CiaRH-dependent control mechanism prevails, csRNA- or HtrA-mediated, depends strongly on the growth conditions.85
In addition to competence control, csRNAs are involved in another CiaRH-dependent phenotype. Mutations in the histidine kinase gene ciaH leading to a hyperactive CiaRH system87,88 have been shown to increase β-lactam resistance. Without csRNAs, these CiaRH hyperactive strains are no longer resistant, but the target(s) involved in this phenotype has not yet been identified. Thus, the csRNAs certainly control at least one more target in S. pneumoniae.
Role of RNA Chaperones in sRNA-Mediated Gene Regulation in Low GC-Gram-Positive Bacteria
An important characteristic of many trans-encoded antisense RNAs from E. coli is their ability to bind the RNA chaperone Hfq (reviewed in ref. 90). Hfq is present in 50% of all sequenced bacterial species, and a few species like Bacillus anthracis encode even two Hfq proteins. Hfq is a homohexamer that is very similar to the eukaryotic Sm proteins involved in splicing.90 It binds to AU-rich sequences in single-stranded regions flanked by one or two stem-loops. Among others, Hfq is involved in mRNA stability, polyadenylation, and translation.90 In Gram-negative bacteria, the majority of trans-encoded sRNAs need Hfq either for their stability or for sRNA/target interaction (reviewed in ref. 10). In 2010, it was shown that sRNAs can displace each other on Hfq on a short time scale by RNA concentration-driven cycling.91
Currently, the only example for Hfq-dependent antisense regulation in Gram-positive bacteria is LhrA from L. monocytogenes (see above).68,69 For two trans-encoded sRNAs in Gram-positive bacteria, B. subtilis SR132 and S. aureus RNAIII,92 Hfq does not impact sRNA/target interaction. In the latter case, this was even tested in three virulent genetic backgrounds.92 However, as hfq expression levels differ between strains, the role of Hfq in S. aureus is discussed controversially (e.g. ref. 93). By contrast, for the trans-encoded sRNA FsrA from B. subtilis, a role of other small putative RNA binding proteins that might act as RNA chaperones—in particular FbpB—has been proposed.16,26 However, it has not yet been demonstrated experimentally that FbpA, B, or C indeed bind RNA. Interestingly, Streptococcus species do not encode Hfq. A study on the interchangeability of Hfq-like proteins between Gram-negative and Gram-positive bacteria demonstrated that neither S. aureus nor Borrelia Hfq expressed chromosomally in S. enterica Typhimurium from the location of endogenous Hfq could functionally substitute Salmonella Hfq in sRNA-mediated regulation and protection from degradation.94 Future research will show whether the LhrA case is an exception and whether other, yet-unidentified RNA binding proteins function as additional RNA chaperones in Gram-positive bacteria. In addition to the Fbp proteins, the SMc01113 protein could be a possible candidate, as it alters sRNA/target mRNA accumulation in Sinorhizobius meliloti,95 but is highly conserved and present in almost all bacteria, also those that lack Hfq.
Future Perspectives
In the near future, the multitude of newly discovered sRNAs in low-GC Gram-positive bacteria will be investigated in detail to identify their targets, to analyze their biological role, and to elucidate their mechanisms of action. It can be expected that novel mechanisms or such known so far only from plasmid-encoded antisense RNAs or from sRNAs in Gram-negative bacteria, will be found for these sRNAs. Likewise, one sRNA might act in cis on one target and in trans on one or several others, thereby employing different modes of action.
As only in one case, LhrA from Listeria monocytogenes, a role for Hfq has been established, it might well be that other chaperones will be detected that play equivalent roles in Gram-positive bacteria. First possible candidates are the small basic proteins FbpA, FbpB, and FbpC, which were linked to the function of B. subtilis FsrA (see above).
Like in E. coli or Salmonella, target mRNAs will be found that are regulated by different sRNAs. In this context, global regulatory networks will be uncovered that implicate both sRNAs and transcriptional repressors and activators.
Furthermore, the small number of dual-function sRNAs will increase considerably, and new unprecedented functions for small peptides encoded by sRNAs will be detected.
The excludon concept recently established for long antisense RNAs (lasRNAs) from Listeria might be confirmed for novel lasRNAs from other low-GC-Gram-positive bacteria. The modes of action used by the lasRNAs may not only comprise classical antisense RNA concepts or RNA interference, but so far unanticipated mechanisms. Additionally, sRNAs might be found that act directly on the genome like e.g., the siRNAs from Schizosaccharomyces pombe that are involved in chromatin silencing (reviewed in ref. 96).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Acknowledgments
This study was supported by grants BR1552/6-1 to 6-3 and by grants BR1552/7-1 and 7-2 of the priority program SPP1258 from the Deutsche Forschungsgemeinschaft to Brantl S and BR974/5-1 of SPP1258 to Brückner R.
References
- 1.Brantl S. Antisense RNAs in plasmids: control of replication and maintenance. Plasmid. 2002;48:165–73. doi: 10.1016/S0147-619X(02)00108-7. [DOI] [PubMed] [Google Scholar]
- 2.Rasmussen S, Nielsen HB, Jarmer H. The transcriptionally active regions in the genome of Bacillus subtilis. Mol Microbiol. 2009;73:1043–57. doi: 10.1111/j.1365-2958.2009.06830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Irnov I, Sharma CM, Vogel J, Winkler WC. Identification of regulatory RNAs in Bacillus subtilis. Nucleic Acids Res. 2010;38:6637–51. doi: 10.1093/nar/gkq454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wiegand S, Dietrich S, Hertel R, Bongaerts J, Evers S, Volland S, Daniel R, Liesegang H. RNA-Seq of Bacillus licheniformis: active regulatory RNA features expressed within a productive fermentation. BMC Genomics. 2013;14:667. doi: 10.1186/1471-2164-14-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fouquier d’Hérouel A, Wessner F, Halpern D, Ly-Vu J, Kennedy SP, Serror P, Aurell E, Repoila F. A simple and efficient method to search for selected primary transcripts: non-coding and antisense RNAs in the human pathogen Enterococcus faecalis. Nucleic Acids Res. 2011;39:e46. doi: 10.1093/nar/gkr012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shioya K, Michaux C, Kuenne C, Hain T, Verneuil N, Budin-Verneuil A, Hartsch T, Hartke A, Giard J-C. Genome-wide identification of small RNAs in the opportunistic pathogen Enterococcus faecalis V583. PLoS One. 2011;6:e23948. doi: 10.1371/journal.pone.0023948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Soutourina OA, Monot M, Boudry P, Saujet L, Pichon C, Sismeiro O, Semenova E, Severinov K, Le Bouguenec C, Coppée JY, et al. Genome-wide identification of regulatory RNAs in the human pathogen Clostridium difficile. PLoS Genet. 2013;9:e1003493. doi: 10.1371/journal.pgen.1003493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tan Y, Liu J, Chen X, Zheng H, Li F. RNA-seq-based comparative transcriptome analysis of the syngas-utilizing bacterium Clostridium ljungdahlii DSM 13528 grown autotrophically and heterotrophically. Mol Biosyst. 2013;9:2775–84. doi: 10.1039/c3mb70232d. [DOI] [PubMed] [Google Scholar]
- 9.Venkataramanan KP, Jones SW, McCormick KP, Kunjeti SG, Ralston MT, Meyers BC, Papoutsakis ET. The Clostridium small RNome that responds to stress: the paradigm and importance of toxic metabolite stress in C. acetobutylicum. BMC Genomics. 2013;14:849. doi: 10.1186/1471-2164-14-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brantl S. Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol. 2009;4:85–103. doi: 10.2217/17460913.4.1.85. [DOI] [PubMed] [Google Scholar]
- 11.Darfeuille F, Unoson C, Vogel J, Wagner EG. An antisense RNA inhibits translation by competing with standby ribosomes. Mol Cell. 2007;26:381–92. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 12.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–7. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brantl S. Acting antisense: plasmid- and chromosome-encoded sRNAs from Gram-positive bacteria. Future Microbiol. 2012;7:853–71. doi: 10.2217/fmb.12.59. [DOI] [PubMed] [Google Scholar]
- 14.Romby P, Charpentier E. An overview of RNAs with regulatory functions in gram-positive bacteria. Cell Mol Life Sci. 2010;67:217–37. doi: 10.1007/s00018-009-0162-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Licht A, Preis S, Brantl S. Implication of CcpN in the regulation of a novel untranslated RNA (SR1) in Bacillus subtilis. Mol Microbiol. 2005;58:189–206. doi: 10.1111/j.1365-2958.2005.04810.x. [DOI] [PubMed] [Google Scholar]
- 16.Gaballa A, Antelmann H, Aguilar C, Khakh S-K, Song K-B, Smaldone GT, Helmann JD. The Bacillus subtilis iron-sparing response is mediated by a Fur-regulated small RNA and three small, basic proteins. Proc Natl Acad Sci U S A. 2008;105:11927–32. doi: 10.1073/pnas.0711752105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geissmann T, Chevalier C, Cros M-J, Boisset S, Fechter P, Noirot C, Schrenzel J, François P, Vandenesch F, Gaspin C, et al. A search for small noncoding RNAs in Staphylococcus aureus reveals a conserved sequence motif for regulation. Nucleic Acids Res. 2009;37:7239–57. doi: 10.1093/nar/gkp668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569–77. doi: 10.1002/j.1460-2075.1995.tb00136.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaito C, Saito Y, Ikuo M, Omae Y, Mao H, Nagano G, Fujiyuki T, Numata S, Han X, Obata K, et al. Mobile genetic element SCCmec-encoded psm-mec RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS Pathog. 2013;9:e1003269. doi: 10.1371/journal.ppat.1003269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mangold M, Siller M, Roppenser B, Vlaminckx BJ, Penfound TA, Klein R, Novak R, Novick RP, Charpentier E. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol Microbiol. 2004;53:1515–27. doi: 10.1111/j.1365-2958.2004.04222.x. [DOI] [PubMed] [Google Scholar]
- 21.Gimpel M, Heidrich N, Mäder U, Krügel H, Brantl S. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Mol Microbiol. 2010;76:990–1009. doi: 10.1111/j.1365-2958.2010.07158.x. [DOI] [PubMed] [Google Scholar]
- 22.Shimizu T, Yaguchi H, Ohtani K, Banu S, Hayashi H. Clostridial VirR/VirS regulon involves a regulatory RNA molecule for expression of toxins. Mol Microbiol. 2002;43:257–65. doi: 10.1046/j.1365-2958.2002.02743.x. [DOI] [PubMed] [Google Scholar]
- 23.Roberts SA, Scott JR. RivR and the small RNA RivX: the missing links between the CovR regulatory cascade and the Mga regulon. Mol Microbiol. 2007;66:1506–22. doi: 10.1111/j.1365-2958.2007.06015.x. [DOI] [PubMed] [Google Scholar]
- 24.Lasa I, Toledo-Arana A, Gingeras TR. An effort to make sense of antisense transcription in bacteria. RNA Biol. 2012;9:1039–44. doi: 10.4161/rna.21167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smaldone GT, Revelles O, Gaballa A, Sauer U, Antelmann H, Helmann JD. A global investigation of the Bacillus subtilis iron-sparing response identifies major changes in metabolism. J Bacteriol. 2012;194:2594–605. doi: 10.1128/JB.05990-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smaldone GT, Antelmann H, Gaballa A, Helmann JD. The FsrA sRNA and FbpB protein mediate the iron-dependent induction of the Bacillus subtilis lutABC iron-sulfur-containing oxidases. J Bacteriol. 2012;194:2586–93. doi: 10.1128/JB.05567-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Preis H, Eckart RA, Gudipati RK, Heidrich N, Brantl S. CodY activates transcription of a small RNA in Bacillus subtilis. J Bacteriol. 2009;191:5446–57. doi: 10.1128/JB.00602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchais A, Duperrier S, Durand S, Gautheret D, Stragier P. CsfG, a sporulation-specific, small non-coding RNA highly conserved in endospore formers. RNA Biol. 2011;8:358–64. doi: 10.4161/rna.8.3.14998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silvaggi JM, Perkins JB, Losick R. Genes for small, noncoding RNAs under sporulation control in Bacillus subtilis. J Bacteriol. 2006;188:532–41. doi: 10.1128/JB.188.2.532-541.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmalisch M, Maiques E, Nikolov L, Camp AH, Chevreux B, Muffler A, Rodriguez S, Perkins J, Losick R. Small genes under sporulation control in the Bacillus subtilis genome. J Bacteriol. 2010;192:5402–12. doi: 10.1128/JB.00534-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heidrich N, Chinali A, Gerth U, Brantl S. The small untranslated RNA SR1 from the Bacillus subtilis genome is involved in the regulation of arginine catabolism. Mol Microbiol. 2006;62:520–36. doi: 10.1111/j.1365-2958.2006.05384.x. [DOI] [PubMed] [Google Scholar]
- 32.Heidrich N, Moll I, Brantl S. In vitro analysis of the interaction between the small RNA SR1 and its primary target ahrC mRNA. Nucleic Acids Res. 2007;35:4331–46. doi: 10.1093/nar/gkm439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Licht A, Brantl S. Transcriptional repressor CcpN from Bacillus subtilis compensates asymmetric contact distribution by cooperative binding. J Mol Biol. 2006;364:434–48. doi: 10.1016/j.jmb.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 34.Licht A, Golbik R, Brantl S. Identification of ligands affecting the activity of the transcriptional repressor CcpN from Bacillus subtilis. J Mol Biol. 2008;380:17–30. doi: 10.1016/j.jmb.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 35.Licht A, Brantl S. The transcriptional repressor CcpN from Bacillus subtilis uses different repression mechanisms at different promoters. J Biol Chem. 2009;284:30032–8. doi: 10.1074/jbc.M109.033076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gimpel M, Preis H, Barth E, Gramzow L, Brantl S. SR1--a small RNA with two remarkably conserved functions. Nucleic Acids Res. 2012;40:11659–72. doi: 10.1093/nar/gks895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gimpel M, Brantl S. Construction of a modular plasmid family for chromosomal integration in Bacillus subtilis. J Microbiol Methods. 2012;91:312–7. doi: 10.1016/j.mimet.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 38.Brantl S. Bacterial type I toxin-antitoxin systems. RNA Biol. 2012;9:1488–90. doi: 10.4161/rna.23045. [DOI] [PubMed] [Google Scholar]
- 39.Silvaggi JM, Perkins JB, Losick R. Small untranslated RNA antitoxin in Bacillus subtilis. J Bacteriol. 2005;187:6641–50. doi: 10.1128/JB.187.19.6641-6650.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jahn N, Preis H, Wiedemann C, Brantl S. BsrG/SR4 from Bacillus subtilis--the first temperature-dependent type I toxin-antitoxin system. Mol Microbiol. 2012;83:579–98. doi: 10.1111/j.1365-2958.2011.07952.x. [DOI] [PubMed] [Google Scholar]
- 41.Durand S, Jahn N, Condon C, Brantl S. Type I toxin-antitoxin systems in Bacillus subtilis. RNA Biol. 2012;9:1491–7. doi: 10.4161/rna.22358. [DOI] [PubMed] [Google Scholar]
- 42.Durand S, Gilet L, Condon C. The essential function of B. subtilis RNase III is to silence foreign toxin genes. PLoS Genet. 2012;8:e1003181. doi: 10.1371/journal.pgen.1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jahn N, Brantl S. One antitoxin--two functions: SR4 controls toxin mRNA decay and translation. Nucleic Acids Res. 2013;41:9870–80. doi: 10.1093/nar/gkt735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr Opin Microbiol. 2007;10:102–9. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 45.Eiamphungporn W, Helmann JD. Extracytoplasmic function sigma factors regulate expression of the Bacillus subtilis yabE gene via a cis-acting antisense RNA. J Bacteriol. 2009;191:1101–5. doi: 10.1128/JB.01530-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benito Y, Kolb FA, Romby P, Lina G, Etienne J, Vandenesch F. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA. 2000;6:668–79. doi: 10.1017/S1355838200992550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, et al. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353–66. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol. 2006;61:1038–48. doi: 10.1111/j.1365-2958.2006.05292.x. [DOI] [PubMed] [Google Scholar]
- 49.Chevalier C, Boisset S, Romilly C, Masquida B, Fechter P, Geissmann T, Vandenesch F, Romby P. Staphylococcus aureus RNAIII binds to two distant regions of coa mRNA to arrest translation and promote mRNA degradation. PLoS Pathog. 2010;6:e1000809. doi: 10.1371/journal.ppat.1000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–35. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xue T, Zhang X, Sun H, Sun B. ArtR, a novel sRNA of Staphylococcus aureus, regulates α-toxin expression by targeting the 5′ UTR of sarT mRNA. Med Microbiol Immunol. 2013 doi: 10.1007/s00430-013-0307-0. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 52.Pichon C, Felden B. Small RNA genes expressed from Staphylococcus aureus genomic and pathogenicity islands with specific expression among pathogenic strains. Proc Natl Acad Sci U S A. 2005;102:14249–54. doi: 10.1073/pnas.0503838102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sayed N, Nonin-Lecomte S, Réty S, Felden B. Functional and structural insights of a Staphylococcus aureus apoptotic-like membrane peptide from a toxin-antitoxin module. J Biol Chem. 2012;287:43454–63. doi: 10.1074/jbc.M112.402693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sayed N, Jousselin A, Felden B. A cis-antisense RNA acts in trans in Staphylococcus aureus to control translation of a human cytolytic peptide. Nat Struct Mol Biol. 2012;19:105–12. doi: 10.1038/nsmb.2193. [DOI] [PubMed] [Google Scholar]
- 55.Chabelskaya S, Gaillot O, Felden B. A Staphylococcus aureus small RNA is required for bacterial virulence and regulates the expression of an immune-evasion molecule. PLoS Pathog. 2010;6:e1000927. doi: 10.1371/journal.ppat.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bohn C, Rigoulay C, Chabelskaya S, Sharma CM, Marchais A, Skorski P, Borezée-Durant E, Barbet R, Jacquet E, Jacq A, et al. Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010;38:6620–36. doi: 10.1093/nar/gkq462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lasa I, Toledo-Arana A, Dobin A, Villanueva M, de los Mozos IR, Vergara-Irigaray M, Segura V, Fagegaltier D, Penadés JR, Valle J, et al. Genome-wide antisense transcription drives mRNA processing in bacteria. Proc Natl Acad Sci U S A. 2011;108:20172–7. doi: 10.1073/pnas.1113521108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morrison JM, Miller EW, Benson MA, Alonzo F, 3rd, Yoong P, Torres VJ, Hinrichs SH, Dunman PM. Characterization of SSR42, a novel virulence factor regulatory RNA that contributes to the pathogenesis of a Staphylococcus aureus USA300 representative. J Bacteriol. 2012;194:2924–38. doi: 10.1128/JB.06708-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tomasini A, François P, Howden BP, Fechter P, Romby P, Caldelari I. The importance of regulatory RNAs in Staphylococcus aureus. Infect Genet Evol. 2013 doi: 10.1016/j.meegid.2013.11.016. Forthcoming. [DOI] [PubMed] [Google Scholar]
- 60.Janssen PJ, Jones DT, Woods DR. Studies on Clostridium acetobutylicum glnA promoters and antisense RNA. Mol Microbiol. 1990;4:1575–83. doi: 10.1111/j.1365-2958.1990.tb02069.x. [DOI] [PubMed] [Google Scholar]
- 61.Fierro-Monti IP, Reid SJ, Woods DR. Differential expression of a Clostridium acetobutylicum antisense RNA: implications for regulation of glutamine synthetase. J Bacteriol. 1992;174:7642–7. doi: 10.1128/jb.174.23.7642-7647.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.André G, Even S, Putzer H, Burguière P, Croux C, Danchin A, Martin-Verstraete I, Soutourina O. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 2008;36:5955–69. doi: 10.1093/nar/gkn601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okumura K, Ohtani K, Hayashi H, Shimizu T. Characterization of genes regulated directly by the VirR/VirS system in Clostridium perfringens. J Bacteriol. 2008;190:7719–27. doi: 10.1128/JB.01573-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Obana N, Nakamura K. A novel toxin regulator, the CPE1446-CPE1447 protein heteromeric complex, controls toxin genes in Clostridium perfringens. J Bacteriol. 2011;193:4417–24. doi: 10.1128/JB.00262-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohtani K, Bhowmik SK, Hayashi H, Shimizu T. Identification of a novel locus that regulates expression of toxin genes in Clostridium perfringens. FEMS Microbiol Lett. 2002;209:113–8. doi: 10.1111/j.1574-6968.2002.tb11118.x. [DOI] [PubMed] [Google Scholar]
- 66.Ohtani K, Hirakawa H, Paredes-Sabja D, Tashiro K, Kuhara S, Sarker MR, Shimizu T. Unique regulatory mechanism of sporulation and enterotoxin production in Clostridium perfringens. J Bacteriol. 2013;195:2931–6. doi: 10.1128/JB.02152-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Y, Indurthi DC, Jones SW, Papoutsakis ET. Small RNAs in the genus Clostridium. MBio. 2011;2:e00340–10. doi: 10.1128/mBio.00340-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Christiansen JK, Nielsen JS, Ebersbach T, Valentin-Hansen P, Søgaard-Andersen L, Kallipolitis BH. Identification of small Hfq-binding RNAs in Listeria monocytogenes. RNA. 2006;12:1383–96. doi: 10.1261/rna.49706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nielsen JS, Lei LK, Ebersbach T, Olsen AS, Klitgaard JK, Valentin-Hansen P, Kallipolitis BH. Defining a role for Hfq in Gram-positive bacteria: evidence for Hfq-dependent antisense regulation in Listeria monocytogenes. Nucleic Acids Res. 2010;38:907–19. doi: 10.1093/nar/gkp1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nielsen JS, Larsen MH, Lillebæk EM, Bergholz TM, Christiansen MH, Boor KJ, Wiedmann M, Kallipolitis BH. A small RNA controls expression of the chitinase ChiA in Listeria monocytogenes. PLoS One. 2011;6:e19019. doi: 10.1371/journal.pone.0019019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Toledo-Arana A, Dussurget O, Nikitas G, Sesto N, Guet-Revillet H, Balestrino D, Loh E, Gripenland J, Tiensuu T, Vaitkevicius K, et al. The Listeria transcriptional landscape from saprophytism to virulence. Nature. 2009;459:950–6. doi: 10.1038/nature08080. [DOI] [PubMed] [Google Scholar]
- 72.Wurtzel O, Sesto N, Mellin JR, Karunker I, Edelheit S, Bécavin C, Archambaud C, Cossart P, Sorek R. Comparative transcriptomics of pathogenic and non-pathogenic Listeria species. Mol Syst Biol. 2012;8:583. doi: 10.1038/msb.2012.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sesto N, Wurtzel O, Archambaud C, Sorek R, Cossart P. The excludon: a new concept in bacterial antisense RNA-mediated gene regulation. Nat Rev Microbiol. 2013;11:75–82. doi: 10.1038/nrmicro2934. [DOI] [PubMed] [Google Scholar]
- 74.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–9. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 75.Ramirez-Peña E, Treviño J, Liu Z, Perez N, Sumby P. The group A Streptococcus small regulatory RNA FasX enhances streptokinase activity by increasing the stability of the ska mRNA transcript. Mol Microbiol. 2010;78:1332–47. doi: 10.1111/j.1365-2958.2010.07427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Steiner K, Malke H. Dual control of streptokinase and streptolysin S production by the covRS and fasCAX two-component regulators in Streptococcus dysgalactiae subsp. equisimilis. Infect Immun. 2002;70:3627–36. doi: 10.1128/IAI.70.7.3627-3636.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Le Rhun A, Charpentier E. Small RNAs in streptococci. RNA Biol. 2012;9:414–26. doi: 10.4161/rna.20104. [DOI] [PubMed] [Google Scholar]
- 78.Acebo P, Martin-Galiano AJ, Navarro S, Zaballos A, Amblar M. Identification of 88 regulatory small RNAs in the TIGR4 strain of the human pathogen Streptococcus pneumoniae. RNA. 2012;18:530–46. doi: 10.1261/rna.027359.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Håvarstein LS, Gaustad P, Nes IF, Morrison DA. Identification of the streptococcal competence-pheromone receptor. Mol Microbiol. 1996;21:863–9. doi: 10.1046/j.1365-2958.1996.521416.x. [DOI] [PubMed] [Google Scholar]
- 80.Mann B, van Opijnen T, Wang J, Obert C, Wang YD, Carter R, McGoldrick DJ, Ridout G, Camilli A, Tuomanen EI, et al. Control of virulence by small RNAs in Streptococcus pneumoniae. PLoS Pathog. 2012;8:e1002788. doi: 10.1371/journal.ppat.1002788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Halfmann A, Kovács M, Hakenbeck R, Brückner R. Identification of the genes directly controlled by the response regulator CiaR in Streptococcus pneumoniae: five out of 15 promoters drive expression of small non-coding RNAs. Mol Microbiol. 2007;66:110–26. doi: 10.1111/j.1365-2958.2007.05900.x. [DOI] [PubMed] [Google Scholar]
- 82.Tsui H-CT, Mukherjee D, Ray VA, Sham LT, Feig AL, Winkler ME. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J Bacteriol. 2010;192:264–79. doi: 10.1128/JB.01204-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Marx P, Nuhn M, Kovács M, Hakenbeck R, Brückner R. Identification of genes for small non-coding RNAs that belong to the regulon of the two-component regulatory system CiaRH in Streptococcus. BMC Genomics. 2010;11:661. doi: 10.1186/1471-2164-11-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Perez N, Treviño J, Liu Z, Ho SC, Babitzke P, Sumby P. A genome-wide analysis of small regulatory RNAs in the human pathogen group A Streptococcus. PLoS One. 2009;4:e7668. doi: 10.1371/journal.pone.0007668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schnorpfeil A, Kranz M, Kovács M, Kirsch C, Gartmann J, Brunner I, Bittmann S, Brückner R. Target evaluation of the non-coding csRNAs reveals a link of the two-component regulatory system CiaRH to competence control in Streptococcus pneumoniae R6. Mol Microbiol. 2013;89:334–49. doi: 10.1111/mmi.12277. [DOI] [PubMed] [Google Scholar]
- 86.Håvarstein LS, Coomaraswamy G, Morrison DA. An unmodified heptadecapeptide pheromone induces competence for genetic transformation in Streptococcus pneumoniae. Proc Natl Acad Sci U S A. 1995;92:11140–4. doi: 10.1073/pnas.92.24.11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guenzi E, Gasc AM, Sicard MA, Hakenbeck R. A two-component signal-transducing system is involved in competence and penicillin susceptibility in laboratory mutants of Streptococcus pneumoniae. Mol Microbiol. 1994;12:505–15. doi: 10.1111/j.1365-2958.1994.tb01038.x. [DOI] [PubMed] [Google Scholar]
- 88.Müller M, Marx P, Hakenbeck R, Brückner R. Effect of new alleles of the histidine kinase gene ciaH on the activity of the response regulator CiaR in Streptococcus pneumoniae R6. Microbiology. 2011;157:3104–12. doi: 10.1099/mic.0.053157-0. [DOI] [PubMed] [Google Scholar]
- 89.Cassone M, Gagne AL, Spruce LA, Seeholzer SH, Sebert ME. The HtrA protease from Streptococcus pneumoniae digests both denatured proteins and the competence-stimulating peptide. J Biol Chem. 2012;287:38449–59. doi: 10.1074/jbc.M112.391482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat Rev Microbiol. 2011;9:578–89. doi: 10.1038/nrmicro2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fender A, Elf J, Hampel K, Zimmermann B, Wagner EGH. RNAs actively cycle on the Sm-like protein Hfq. Genes Dev. 2010;24:2621–6. doi: 10.1101/gad.591310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bohn C, Rigoulay C, Bouloc P. No detectable effect of RNA-binding protein Hfq absence in Staphylococcus aureus. BMC Microbiol. 2007;7:10. doi: 10.1186/1471-2180-7-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Liu Y, Wu N, Dong J, Gao Y, Zhang X, Mu C, Shao N, Yang G. Hfq is a global regulator that controls the pathogenicity of Staphylococcus aureus. PLoS One. 2010;5:e13069. doi: 10.1371/journal.pone.0013069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Rochat T, Bouloc P, Yang Q, Bossi L, Figueroa-Bossi N. Lack of interchangeability of Hfq-like proteins. Biochimie. 2012;94:1554–9. doi: 10.1016/j.biochi.2012.01.016. [DOI] [PubMed] [Google Scholar]
- 95.Pandey SP, Minesinger BK, Kumar J, Walker GC. A highly conserved protein of unknown function in Sinorhizobium meliloti affects sRNA regulation similar to Hfq. Nucleic Acids Res. 2011;39:4691–708. doi: 10.1093/nar/gkr060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grewal SI. RNAi-dependent formation of heterochromatin and its diverse functions. Curr Opin Genet Dev. 2010;20:134–41. doi: 10.1016/j.gde.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Lee JM, Zhang S, Saha S, Santa Anna S, Jiang C, Perkins J. RNA expression analysis using an antisense Bacillus subtilis genome array. J Bacteriol. 2001;183:7371–80. doi: 10.1128/JB.183.24.7371-7380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Saito S, Kakeshita H, Nakamura K. Novel small RNA-encoding genes in the intergenic regions of Bacillus subtilis. Gene. 2009;428:2–8. doi: 10.1016/j.gene.2008.09.024. [DOI] [PubMed] [Google Scholar]
- 99.Nicolas P, Mäder U, Dervyn E, Rochat T, Leduc A, Pigeonneau N, Bidnenko E, Marchadier E, Hoebeke M, Aymerich S, et al. Condition-dependent transcriptome reveals high-level regulatory architecture in Bacillus subtilis. Science. 2012;335:1103–6. doi: 10.1126/science.1206848. [DOI] [PubMed] [Google Scholar]
- 100.Marchais A, Naville M, Bohn C, Bouloc P, Gautheret D. Single-pass classification of all noncoding sequences in a bacterial genome using phylogenetic profiles. Genome Res. 2009;19:1084–92. doi: 10.1101/gr.089714.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Beaume M, Hernandez D, Farinelli L, Deluen C, Linder P, Gaspin C, Romby P, Schrenzel J, François P. Cartography of methicillin-resistant S. aureus transcripts: detection, orientation and temporal expression during growth phase and stress conditions. PLoS One. 2010;5:e10725. doi: 10.1371/journal.pone.0010725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Abu-Qatouseh LF, Chinni SV, Seggewiss J, Proctor RA, Brosius J, Rozhdestvensky TS, Peters G, von Eiff C, Becker K. Identification of differentially expressed small non-protein-coding RNAs in Staphylococcus aureus displaying both the normal and the small-colony variant phenotype. J Mol Med (Berl) 2010;88:565–75. doi: 10.1007/s00109-010-0597-2. [DOI] [PubMed] [Google Scholar]
- 103.Howden BP, Beaume M, Harrison PF, Hernandez D, Schrenzel J, Seemann T, Francois P, Stinear TP. Analysis of the small RNA transcriptional response in multidrug-resistant Staphylococcus aureus after antimicrobial exposure. Antimicrob Agents Chemother. 2013;57:3864–74. doi: 10.1128/AAC.00263-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in Listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35:962–74. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Oliver HF, Orsi RH, Ponnala L, Keich U, Wang W, Sun Q, Cartinhour SW, Filiatrault MJ, Wiedmann M, Boor KJ. Deep RNA sequencing of L. monocytogenes reveals overlapping and extensive stationary phase and sigma B-dependent transcriptomes, including multiple highly transcribed noncoding RNAs. BMC Genomics. 2009;10:641. doi: 10.1186/1471-2164-10-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mraheil MA, Billion A, Mohamed W, Mukherjee K, Kuenne C, Pischimarov J, Krawitz C, Retey J, Hartsch T, Chakraborty T, et al. The intracellular sRNA transcriptome of Listeria monocytogenes during growth in macrophages. Nucleic Acids Res. 2011;39:4235–48. doi: 10.1093/nar/gkr033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mellin JR, Cossart P. The non-coding RNA world of the bacterial pathogen Listeria monocytogenes. RNA Biol. 2012;9:372–8. doi: 10.4161/rna.19235. [DOI] [PubMed] [Google Scholar]
- 108.Patenge N, Billion A, Raasch P, Normann J, Wisniewska-Kucper A, Retey J, Boisguérin V, Hartsch T, Hain T, Kreikemeyer B. Identification of novel growth phase- and media-dependent small non-coding RNAs in Streptococcus pyogenes M49 using intergenic tiling arrays. BMC Genomics. 2012;13:550. doi: 10.1186/1471-2164-13-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Tesorero RA, Yu N, Wright JO, Svencionis JP, Cheng Q, Kim JH, Cho KH. Novel regulatory small RNAs in Streptococcus pyogenes. PLoS One. 2013;8:e64021. doi: 10.1371/journal.pone.0064021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kumar R, Shah P, Swiatlo E, Burgess SC, Lawrence ML, Nanduri B. Identification of novel non-coding small RNAs from Streptococcus pneumoniae TIGR4 using high-resolution genome tiling arrays. BMC Genomics. 2010;11:350. doi: 10.1186/1471-2164-11-350. [DOI] [PMC free article] [PubMed] [Google Scholar]