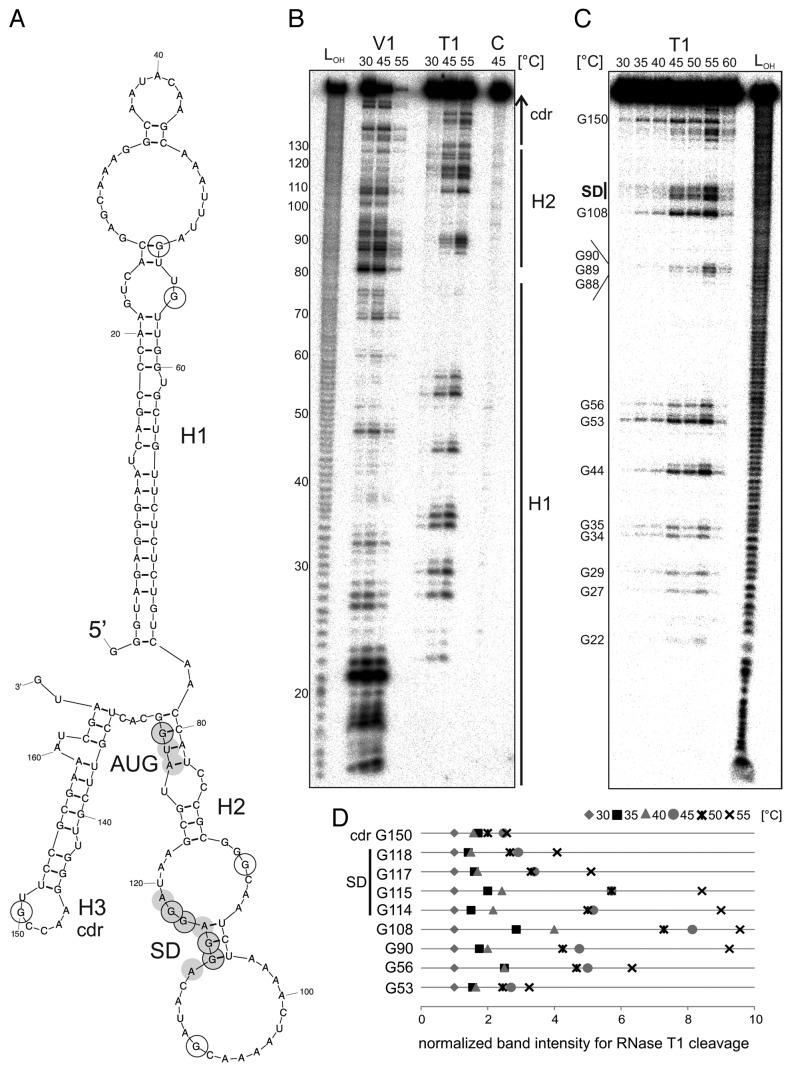
Figure 8. Temperature-dependent structural alterations of the hspA-5′UTR. (A) Model of the T. elongatus hspA-5′UTR and 40 Nt coding region as well as the first two Nt of the blunt end restriction site HpyCH4v according to a secondary structure prediction computed with mfold.86 The structure exhibits two RNA hairpins H1 and H2 in the 5′UTR, the second containing SD region and AUG start codon, as well as a third H3 in the coding region. SD sequence und AUG start codon are highlighted by gray circles. Black encircled Nt are referred to in the quantification in (D). (B) Enzymatic structure probing of the hspA-5′UTR with 40 Nt coding region. In vitro transcribed RNA was 5′labeled81 and subjected to partial digestion82 with RNase V1 (0.001U) and T1 (0.002U) at the indicated temperatures. For the control, (C) A. dest was added instead of enzyme. An alkaline ladder (LOH) was loaded for orientation. Samples were separated on an 8% denaturing polyacrylamide gel. (C) Temperature stability of the hspA RNA structure. Labeled RNA was digested by RNase T1 (0.002U) within a temperature range from 30–60 °C in 5 degree steps. The probing reveals slight accessibility of the SD region for RNase T1 at 45 °C and high susceptibility at 55 °C. Please note that RNase T1 was inactive at 60 °C. (D) Quantification of altered RNase T1 cleavage intensity between 30–55 °C for selected guanines. Band intensities were quantified from (C) using the Alpha Ease software (Alpha Innotech, Biozym). Shown are relative intensities for each Nt position with band intensity at 30 °C set as 1. cdr, coding region.

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.